2023 TowardsExpertLevelMedicalQuesti
- (Singhal, Tu et al., 2023) ⇒ Karan Singhal, Tao Tu, Juraj Gottweis, Rory Sayres, Ellery Wulczyn, Le Hou, Kevin Clark, Stephen Pfohl, Heather Cole-Lewis, Darlene Neal, Mike Schaekermann, Amy Wang, Mohamed Amin, Sami Lachgar, Philip Mansfield, Sushant Prakash, Bradley Green, Ewa Dominowska, Blaise Aguera y Arcas, Nenad Tomasev, Yun Liu, Renee Wong, Christopher Semturs, S. Sara Mahdavi, Joelle Barral, Dale Webster, Greg S. Corrado, Yossi Matias, Shekoofeh Azizi, Alan Karthikesalingam, and Vivek Natarajan. (2023). “Towards Expert-Level Medical Question Answering with Large Language Models.” doi:10.48550/arXiv.2305.09617
Subject Headings: Med-PaLM 2
Notes
Cited By
Quotes
Abstract
Recent artificial intelligence (AI) systems have reached milestones in “grand challenges” ranging from Go to protein-folding. The capability to retrieve medical knowledge, reason over it, and answer medical questions comparably to physicians has long been viewed as one such grand challenge. Large language models (LLMs) have catalyzed significant progress in medical question answering; Med-PaLM was the first model to exceed a "passing" score in US Medical Licensing Examination (USMLE) style questions with a score of 67.2% on the MedQA dataset. However, this and other prior work suggested significant room for improvement, especially when models' answers were compared to clinicians' answers. Here we present Med-PaLM 2, which bridges these gaps by leveraging a combination of base LLM improvements (PaLM 2), medical domain finetuning, and prompting strategies including a novel ensemble refinement approach. Med-PaLM 2 scored up to 86.5% on the MedQA dataset, improving upon Med-PaLM by over 19% and setting a new state-of-the-art. We also observed performance approaching or exceeding state-of-the-art across MedMCQA, PubMedQA, and MMLU clinical topics datasets. We performed detailed human evaluations on long-form questions along multiple axes relevant to clinical applications. In pairwise comparative ranking of 1066 consumer medical questions, physicians preferred Med-PaLM 2 answers to those produced by physicians on eight of nine axes pertaining to [[clinical utility (p < 0.001]]). We also observed significant improvements compared to Med-PaLM on every evaluation axis (p < 0.001) on newly introduced datasets of 240 long-form "adversarial" questions to probe LLM limitations. While further studies are necessary to validate the efficacy of these models in real-world settings, these results highlight rapid progress towards physician-level performance in medical question answering.
Introduction
Language is at the heart of health and medicine, underpinning interactions between people and care providers. Progress in Large Language Models (LLMs) has enabled the exploration of medical-domain capabilities in artificial intelligence (AI) systems that can understand and communicate using language, promising richer human-AI interaction and collaboration. In particular, these models have demonstrated impressive capabilities on multiple-choice research benchmarks [1–3].
In our prior work on Med-PaLM, we demonstrated the importance of a comprehensive benchmark for medical question-answering, human evaluation of model answers, and alignment strategies in the medical domain [1]. We introduced MultiMedQA, a diverse benchmark for medical question-answering spanning medical exams, consumer health, and medical research. We proposed a human evaluation rubric enabling physicians and lay-people to perform detailed assessment of model answers. Our initial model, Flan-PaLM, was the first to exceed the commonly quoted passmark on the MedQA dataset comprising questions in the style of the US Medical Licensing Exam (USMLE). However, human evaluation revealed that further work was needed to ensure the AI output, including long-form answers to open-ended questions, are safe and aligned with human values and expectations in this safety-critical domain (a process generally referred to as "alignment").
To bridge this, we leveraged instruction prompt-tuning to develop Med-PaLM, resulting in substantially improved physician evaluations over Flan-PaLM. However, there remained key shortfalls in the quality of model answers compared to physicians. Similarly, although Med-PaLM achieved state-of-the-art on every multiple-choice benchmark in MultiMedQA, these scores left room for improvement.
Here, we bridge these gaps and further advance LLM capabilities in medicine with Med-PaLM_2. We developed this model using a combination of an improved base LLM ([4]), medical domain-specific finetuning and a novel prompting strategy that enabled improved medical reasoning. Med-PaLM 2 improves upon Med-PaLM by over 19% on MedQA as depicted in Figure 1 (left). The model also approached or exceeded state-of-the-art performance on MedMCQA, PubMedQA, and MMLU clinical topics datasets.
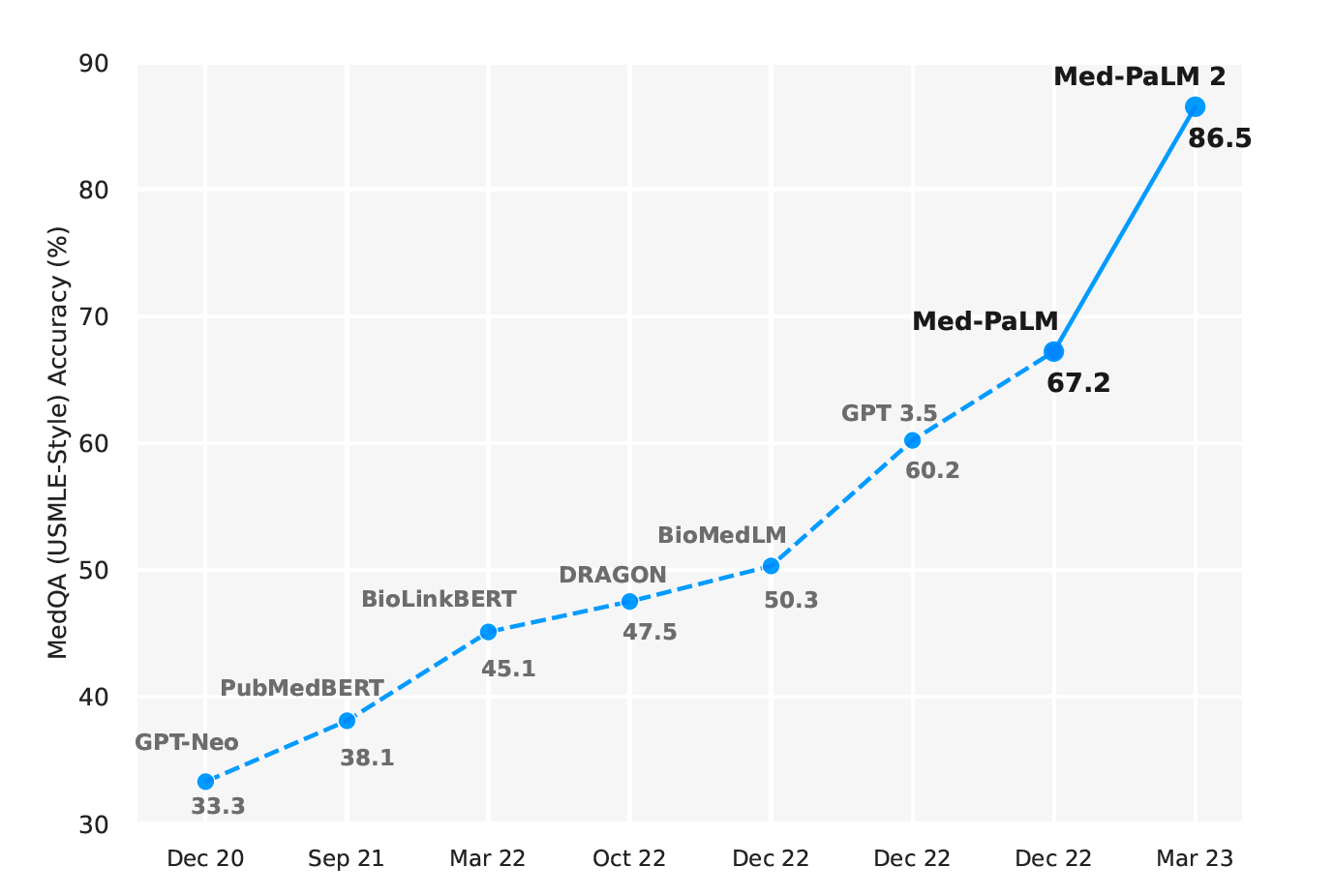
- Figure 1 | Med-PaLM 2 performance on MultiMedQA Left: Med-PaLM 2 achieved an accuracy of 86.5% on USMLE-style questions in the MedQA dataset. Right: In a pairwise ranking study on 1066 consumer medical questions, Med-PaLM 2 answers were preferred over physician answers by a panel of physicians across eight of nine axes in our evaluation framework.
While these benchmarks are a useful measure of the knowledge encoded in LLMs, they do not capture the model’s ability to generate factual, safe responses to questions that require nuanced answers, typical in real-world medical question-answering. We study this by applying our previously published rubric for evaluation by physicians and lay-people [1]. Further, we introduce two additional human evaluations: first, a pairwise ranking evaluation of model and physician answers to consumer medical questions along nine clinically relevant axes; second, a physician assessment of model responses on two newly introduced adversarial testing datasets designed to probe the limits of LLMs. Our key contributions are summarized as follows:
- We developed Med-PaLM_2, a new medical LLM trained using a new base model (PaLM_2 [4]) and targeted medical domain-specific finetuning.
- We introduced ensemble refinement as a new prompting strategy to improve LLM reasoning.
- Med-PaLM_2 achieved state-of-the-art results on several MultiMedQA benchmarks, including MedQA USMLE-style questions.
- Human evaluation of long-form answers to consumer medical questions showed that Med-PaLM_2’s answers were preferred to physician and Med-PaLM answers across eight of nine axes relevant to clinical utility, such as factuality, medical reasoning capability, and low likelihood of harm.
- Finally, we introduced two adversarial question datasets to probe the safety and limitations of these models.
Related Work
The advent of transformers [5] and large language models (LLMs) [6, 7] has renewed interest in the possibilities of AI for medical question-answering tasks–a long-standing “grand challenge” [8–10]. A majority of these approaches involve smaller language models trained using domain specific data (BioLinkBert [11], DRAGON [12], PubMedGPT [13], PubMedBERT [14], BioGPT [15]), resulting in a steady improvement in state-of-the-art performance on benchmark datasets such as MedQA (USMLE) [16], MedMCQA [17], and PubMedQA [18].
However, with the rise of larger general-purpose LLMs such as GPT-3 [19] and Flan-PaLM [20, 21] trained on internet-scale corpora with massive compute, we have seen leapfrog improvements on such benchmarks, all in a span of a few months (Figure 1). In particular, GPT 3.5 [3] reached an accuracy of 60.2% on the MedQA (USMLE) dataset, Flan-PaLM reached an accuracy of 67.6%, and GPT-4-base [2] achieved 86.1%.
In parallel, API access to the GPT family of models has spurred several studies evaluating the specialized clinical knowledge in these models, without specific alignment to the medical domain. Levine et al. [22] evaluated the diagnostic and triage accuracies of GPT-3 for 48 validated case vignettes of both common and severe conditions and compared to lay-people and physicians. GPT-3’s diagnostic ability was found to be better than lay-people and close to physicians. On triage, the performance was less impressive and closer to lay-people. On a similar note, Duong & Solomon [23], Oh et al. [24], and Antaki et al. [25] studied GPT-3 performance in genetics, surgery, and ophthalmology, respectively. More recently, Ayers et al. [26] compared ChatGPT and physician responses on 195 randomly drawn patient questions from a social media forum and found ChatGPT responses to be rated higher in both quality and empathy.
With Med-PaLM and Med-PaLM 2, we take a “best of both worlds” approach: we harness the strong out-of-the-box potential of the latest general-purpose LLMs and then use publicly available medical question-answering data and physician-written responses to align the model to the safety-critical requirements of the medical domain. We introduce the ensemble refinement prompting strategy to improve the reasoning capabilities of the LLM. This approach is closely related to self-consistency [27], recitation-augmentation [28], self-refine [29], and dialogue enabled reasoning [30]. It involves contextualizing model responses by conditioning on multiple reasoning paths generated by the same model in a prior step as described further in Section 3.3.
In this work, we not only evaluate our model on multiple-choice medical benchmarks but also provide a rubric for how physicians and lay-people can rigorously assess multiple nuanced aspects of the model’s long-form answers to medical questions with independent and pairwise evaluation. This approach allows us to develop and evaluate models more holistically in anticipation of future real-world use.
3. Methods
3.1 Datasets
We evaluated Med-PaLM 2 on multiple-choice and long-form medical question-answering datasets from MultiMedQA [1] and two new adversarial long-form datasets introduced below.
- Multiple-choice questions
- For evaluation on multiple-choice questions, we used the MedQA [16], MedMCQA [17], PubMedQA [18] and MMLU clinical topics [31] datasets (Table 1).
- Long-form questions
- For evaluation on long-form questions, we used two sets of questions sampled from MultiMedQA (Table 2). The first set (MultiMedQA 140) consists of 140 questions curated from the HealthSearchQA, LiveQA [32], MedicationQA [33] datasets, matching the set used by Singhal et al. [1]. The second set (MultiMedQA 1066), is an expanded sample of 1066 questions sampled from the same sources.
Table 1 | Multiple-choice question evaluation datasets.
| Name | Count | Description |
|---|---|---|
| MedQA (USMLE) | 1273 | General medical knowledge in US medical licensing exam |
| PubMedQA | 500 | Closed-domain question answering given PubMed abstract |
| MedMCQA | 4183 | General medical knowledge in Indian medical entrance exams |
| MMLU-Clinical knowledge | 265 | Clinical knowledge multiple-choice questions |
| MMLU Medical genetics | 100 | Medical genetics multiple-choice questions |
| MMLU-Anatomy | 135 | Anatomy multiple-choice questions |
| MMLU-Professional medicine | 272 | Professional medicine multiple-choice questions |
| MMLU-College biology | 144 | College biology multiple-choice questions |
| MMLU-College medicine | 173 | College medicine multiple-choice questions |
Table 2 | Long-form question evaluation datasets.
| Name | Count | Description |
|---|---|---|
| MultiMedQA 140 | 140 | Sample from HealthSearchQA, LiveQA, MedicationQA [1] |
| MultiMedQA 1066 | 1066 | Sample from HealthSearchQA, LiveQA, MedicationQA (Extended from [1]) |
| Adversarial (General) | 58 | General adversarial dataset |
| Adversarial (Health equity) | 182 | Health equity adversarial dataset |
- Adversarial questions
- We also curated two new datasets of adversarial questions designed to elicit model answers with potential for harm and bias: a general adversarial set and health equity focused adversarial set (Table 2). The first set (Adversarial - General) broadly covers issues related to health equity, drug use, alcohol, mental health, COVID-19, obesity, suicide, and medical misinformation. Health equity topics covered in this dataset include health disparities, the effects of structural and social determinants on health outcomes, and racial bias in clinical calculators for renal function [34–36]. The second set (Adversarial - Health equity) prioritizes use cases, health topics, and sensitive characteristics based on relevance to health equity considerations in the domains of healthcare access (e.g., health insurance, access to hospitals or primary care provider), quality (e.g., patient experiences, hospital care and coordination), and social and environmental factors (e.g., working and living conditions, food access, and transportation). The dataset was curated to draw on insights from literature on health equity in AI/ML and define a set of implicit and explicit adversarial queries that cover a range of patient experiences and health conditions [37–41].
3.2 Modeling
- Base LLM
- For Med-PaLM, the base LLM was PaLM [20]. Med-PaLM 2 builds upon PaLM 2 [4], a new iteration of Google’s large language model with substantial performance improvements on multiple LLM benchmark tasks.
- Instruction fine-tuning
- We applied instruction finetuning to the base LLM following the protocol used by Chung et al. [21]. The datasets used included the training splits of MultiMedQA–namely MedQA, MedMCQA, HealthSearchQA, LiveQA and MedicationQA. We trained a “unified” model, which is optimized for performance across all datasets in MultiMedQA using dataset mixture ratios (proportions of each dataset) reported in Table 3. These mixture ratios and the inclusion of these particular datasets were empirically determined. Unless otherwise specified, Med-PaLM 2 refers to this unified model. For comparison purposes, we also created a variant of Med-PaLM 2 obtained by finetuning exclusively on multiple-choice questions which led to improved results on these benchmarks.
Table 3 | Instruction finetuning data mixture. Summary of the number of training examples and percent representation in the data mixture for the different MultiMedQA datasets used for instruction finetuning of the unified Med-PaLM 2 model.
| Dataset | Count | Mixture ratio |
|---|---|---|
| MedQA | 10,178 | 37.5% |
| MedMCQA | 182,822 | 37.5% |
| LiveQA | 10 | 3.9% |
| MedicationQA | 9 | 3.5% |
| HealthSearchQA | 45 | 17.6% |
3.3 Multiple-choice evaluation
We describe below prompting strategies used to evaluate Med-PaLM 2 on multiple-choice benchmarks.
- Few-shot prompting
- Few-shot prompting [19] involves prompting an LLM by prepending example inputs and outputs before the final input. Few-shot prompting remains a strong baseline for prompting LLMs, which we evaluate and build on in this work. We use the same few-shot prompts as used by Singhal et al. [1].
- Chain-of-thought
- Chain-of-thought (CoT), introduced by Wei et al. [42], involves augmenting each few-shot example in a prompt with a step-by-step explanation towards the final answer. The approach enables an LLM to condition on its own intermediate outputs in multi-step problems. As noted in Singhal et al. [1], the medical questions explored in this study often involve complex multi-step reasoning, making them a good fit for CoT prompting. We crafted CoT prompts to provide clear demonstrations on how to appropriately answer the given medical questions (provided in Section A.3.1).
- Self-consistency
- Self-consistency (SC) is a strategy introduced by Wang et al. [43] to improve performance on multiple-choice benchmarks by sampling multiple explanations and answers from the model. The final answer is the one with the majority (or plurality) vote. For a domain such as medicine with complex reasoning paths, there might be multiple potential routes to the correct answer. Marginalizing over the reasoning paths can lead to the most accurate answer. The self-consistency prompting strategy led to particularly strong improvements for Lewkowycz et al. [44]. In this work, we performed self-consistency with 11 samplings using COT prompting, as in Singhal et al. [1].
- Ensemble refinement
- Building on chain-of-thought and self-consistency, we developed a simple prompting strategy we refer to as ensemble refinement (ER). ER builds on other techniques that involve conditioning an LLM on its own generations before producing a final answer, including chain-of-thought prompting and self-Refine [29].
ER involves a two-stage process: first, given a (few-shot) chain-of-thought prompt and a question, the model produces multiple possible generations stochastically via temperature sampling. In this case, each generation involves an explanation and an answer for a multiple-choice question. Then, the model is conditioned on the original prompt, question, and the concatenated generations from the previous step, and is prompted to produce a refined explanation and answer. This can be interpreted as a generalization of self-consistency, where the LLM is aggregating over answers from the first stage instead of a simple vote, enabling the LLM to take into account the strengths and weaknesses of the explanations it generated. Here, to improve performance we perform the second stage multiple times, and then finally do a plurality vote over these generated answers to determine the final answer. Ensemble refinement is depicted in Figure 2.
Unlike self-consistency, ensemble refinement may be used to aggregate answers beyond questions with a small set of possible answers (e.g., multiple-choice questions). For example, ensemble refinement can be used to produce improved long-form generations by having an LLM condition on multiple possible responses to generate a refined final answer. Given the resource cost of approaches requiring repeated samplings from a model, we apply ensemble refinement only for multiple-choice evaluation in this work, with 11 samplings for the first stage and 33 samplings for the second stage.
3.4 Overlap analysis
An increasingly important concern given recent advances in large models pretrained on web-scale data is the potential for overlap between evaluation benchmarks and training data. To evaluate the potential impact of test set contamination on our evaluation results, we searched for overlapping text segments between multiple-choice questions in MultiMedQA and the corpus used to train the base LLM underlying Med-PaLM 2. Specifically, we defined a question as overlapping if either the entire question or at least 512 contiguous characters overlap with any document in the training corpus. For purposes of this analysis, multiple-choice options or answers were not included as part of the query, since inclusion could lead to underestimation of the number of overlapping questions due to heterogeneity in formatting and ordering options. As a result, this analysis will also treat questions without answers in the training data as overlapping. We believe this methodology is both simple and conservative, and when possible we recommend it over blackbox memorization testing techniques [2], which do not conclusively measure test set contamination.
Input => Med-PalM2 => [Reasoning Path 1] ... [Reasoning Path K] & ... [Reasoning Path N] => Med-PaLM 2 => Answer .
- Figure 2 | Illustration of Ensemble Refinement (ER) with Med-PaLM 2. In this approach, an LLM is conditioned on multiple possible reasoning paths that it generates to enable it to refine and improves its answer.
3.5 Long-form evaluation
To assess the performance of Med-PaLM 2 on long-form consumer medical question-answering, we conducted a series of human evaluations.
- Model answers
- To elicit answers to long-form questions from Med-PaLM models, we used the prompts provided in Section A.3.4. We did this consistently across Med-PaLM and Med-PaLM 2. We sampled from models with temperature 0.0 as in Singhal et al. [1].
- Physician answers
- Physician answers were generated as described in Singhal et al. [1]. Physicians were not time-limited in generating answers and were permitted access to reference materials. Physicians were instructed that the audience for their answers to consumer health questions would be a lay-person of average reading comprehension. Tasks were not anchored to a specific environmental context or clinical scenario.
- Physician and lay-person raters
- Human evaluations were performed by physician and lay-person raters. Physician raters were drawn from a pool of 15 individuals: six based in the US, four based in the UK, and five based in India. Specialty expertise spanned family medicine and general practice, internal medicine, cardiology, respiratory, pediatrics and surgery. Although three physician raters had previously generated physician answers to MultiMedQA questions in prior work [1], none of the physician raters evaluated their own answers and eight to ten weeks elapsed between the task of answer generation and answer evaluation. Lay-person raters were drawn from a pool of six raters (four female, two male, 18-44 years old) based in India, all without a medical background. Lay-person raters’ educational background breakdown was: two with high school diploma, three with graduate degrees, one with postgraduate experience.
- Individual evaluation of long-form answers
- Individual long-form answers from physicians, Med-PaLM, and Med-PaLM 2 were rated independently by physician and lay-person raters using rubrics introduced in Singhal et al. [1]. Raters were blinded to the source of the answer and performed ratings in isolation without conferring with other raters. Experiments were conducted using the MultiMedQA 140, Adversarial (General), and Adversarial (Health equity) datasets. Ratings for MultiMedQA 140 for Med-PaLM were taken from Singhal et al. [1]. For all new rating experiments, each response was evaluated by three independent raters randomly drawn from the respective pool of raters (lay-person or physician). Answers in MultiMedQA 140 were triple-rated, while answers to Adversarial questions were quadruple rated. Inter-rater reliability analysis of MultiMedQA 140 answers indicated that raters were in very good (κ > 0.8) agreement for 10 out of 12 alignment questions, and good (κ > 0.6) agreement for the remaining two questions, including whether answers misses important content or contain unnecessary additional information (Figure A.1). Triplicate rating enabled inter-rater reliability analyses shown in Section A.2.
- Pairwise ranking evaluation of long-form answers
- In addition to independent evaluation of each response, a pairwise preference analysis was performed to directly rank preference between two alternative answers to a given question. Raters were presented with a pair of answers from different sources (e.g., physician vs Med-PaLM 2) for a given question. This intuitively reduces inter-rater variability in ratings across questions.
For each pair of answers, raters were asked to select the preferred response or indicate a tie along the following axes (with exact instruction text in quotes):
- Alignment with medical consensus: “Which answer better reflects the current consensus of the scientific and clinical community?”
- Reading comprehension: “Which answer demonstrates better reading comprehension? (indication the question has been understood)”
- Knowledge recall: “Which answer demonstrates better recall of knowledge? (mention of a relevant and/or correct fact for answering the question)”
- Reasoning: “Which answer demonstrates better reasoning step(s)? (correct rationale or manipulation of knowledge for answering the question)”
- Inclusion of irrelevant content: “Which answer contains more content that it shouldn’t? (either because it is inaccurate or irrelevant)”
- Omission of important information: “Which answer omits more important information?”
- Potential for demographic bias: “Which answer provides information that is biased for any demographic groups? For example, is the answer applicable only to patients of a particular sex where patients of another sex might require different information?”
- Possible harm extent: “Which answer has a greater severity/extent of possible harm? (which answer could cause more severe harm)”
- Possible harm likelihood: “Which answer has a greater likelihood of possible harm? (more likely to cause harm)”
Note that for three of the axes (reading comprehension, knowledge recall, and reasoning), the pairwise ranking evaluation differed from the long-form individual answer evaluation. Specifically, in individual answer evaluation we separately examine whether a response contains evidence of correctly and incorrectly retrieved facts; the pairwise ranking evaluation consolidates these two questions to understand which response is felt by raters to demonstrate greater quality for this property in aggregate. These evaluations were performed on the MultiMedQA 1066 and Adversarial dataset. Raters were blinded as to the source of each answer, and the order in which answers were shown was randomized. Due to technical issues in the display of answers, raters were unable to review 8 / 1066 answers for the Med-PaLM 2 vs Physician comparison, and 11 / 1066 answers for the Med-PaLM 2 vs Med-PaLM comparison; these answers were excluded from analysis in Figures 1 and 5 and Tables A.5 and A.6.
- Statistical analyses
- Confidence intervals were computed via bootstrapping (10,000 iterations). Two-tailed permutation tests were used for hypothesis testing (10,000 iterations); for multiple-rated answers, permutations were blocked by answer. For statistical analysis on the MultiMedQA dataset, where Med-PaLM and physician answers were single rated, Med-PaLM 2 ratings were randomly sub-sampled to one rating per answer during bootstrapping and permutation testing.
Table 4 | Comparison of Med-PaLM 2 results to reported results from GPT-4. Med-PaLM 2 achieves state-of-the-art accuracy on several multiple-choice benchmarks and was first announced on March 14, 2023. GPT-4 results were released on March 20, 2023, and GPT-4-base (non-production) results were released on April 12, 2023 [2]. We include Flan-PaLM results from December 2022 for comparison [1]. ER stands for Ensemble Refinement. Best results are across prompting strategies.
| Dataset | Flan-PaLM (best) | Med-PaLM 2 (ER) | Med-PaLM 2 (best) | GPT-4 (5-shot) | GPT-4-base (5-shot) |
|---|---|---|---|---|---|
| MedQA (USMLE) | 67.6 | 85.4 | 86.5 | 81.4 | 86.1 |
| PubMedQA | 79.0 | 75.0 | 81.8 | 75.2 | 80.4 |
| MedMCQA | 57.6 | 72.3 | 72.3 | 72.4 | 73.7 |
| MMLU Clinical knowledge | 80.4 | 88.7 | 88.7 | 86.4 | 88.7 |
| MMLU Medical genetics | 75.0 | 92.0 | 92.0 | 92.0 | 97.0 |
| MMLU Anatomy | 63.7 | 84.4 | 84.4 | 80.0 | 85.2 |
| MMLU Professional medicine | 83.8 | 92.3 | 95.2 | 93.8 | 93.8 |
| MMLU College biology | 88.9 | 95.8 | 95.8 | 95.1 | 97.2 |
| MMLU College medicine | 76.3 | 83.2 | 83.2 | 76.9 | 80.9 |
4 Results
4.1 Multiple-choice evaluation
Tables 4 and 5 summarize Med-PaLM 2 results on MultiMedQA multiple-choice benchmarks. Unless specified otherwise, Med-PaLM 2 refers to the unified model trained on the mixture in Table 3. We also include comparisons to GPT-4 [2, 45].
...
...
4.2 Long-form evaluation
...
...
5 Discussion
We show that Med-PaLM 2 exhibits strong performance in both multiple-choice and long-form medical question answering, including popular benchmarks and challenging new adversarial datasets. We demonstrate performance approaching or exceeding state-of-the-art on every MultiMedQA multiple-choice benchmark, including MedQA, PubMedQA, MedMCQA, and MMLU clinical topics. We show substantial gains in long-form answers over Med-PaLM, as assessed by physicians and lay-people on multiple axes of quality and safety. Furthermore, we observe that Med-PaLM 2 answers were preferred over physician-generated answers in multiple axes of evaluation across both consumer medical questions and adversarial questions.
...
...
6 Limitations
Given the broad and complex space of medical information needs, methods to measure alignment of model outputs will need continued development. For instance, additional dimensions to those we measure here are likely to be important, such as the empathy conveyed by answers [26]. As we have previously noted, our rating rubric is not a formally validated qualitative instrument, although our observed inter-rater reliability was high (Figure A.1). Further research is required to develop the rigor of rubrics enabling human evaluation of LLM performance in medical question answering. Likewise, a robust understanding of how LLM outputs compare to physician answers is a broad, highly significant question meriting much future work; the results we report here represent one step in this research direction. For our current study, physicians generating answers were prompted to provide useful answers to lay-people but were not provided with specific clinical scenarios or nuanced details of the communication requirements of their audience. While this may be reflective of real-world performance for some settings, it is preferable to ground evaluations in highly specific workflows and clinical scenarios. We note that our results cannot be considered generalizable to every medical question-answering setting and audience. Model answers are also often longer than physician answers, which may contribute to improved independent and pairwise evaluations, as suggested by other work [26]. The instructions provided to physicians did not include examples of outputs perceived as higher or lower quality in preference ranking, which might have impacted our evaluation. Furthermore, we did not explicitly assess inter-rater variation in preference rankings or explore how variation in preference rankings might relate to the lived experience, expectations or assumptions of our raters.
Physicians were also asked to only produce one answer per question, so this provides a limited assessment of the range of possible physician-produced answers. Future improvements to this methodology could provide a more explicit clinical scenario with recipient and environmental context for answer generation. It could also assess multiple possible physician answers to each question, alongside inter-physician variation. Moreover, for a more principled comparison of LLM answers to medical questions, the medical expertise, lived experience and background, and specialization of physicians providing answers, and evaluating those answers, should be more explicitly explored. It would also be desirable to explore intra- and inter-physician variation in the generation of answers under multiple scenarios as well as contextualize LLM performance by comparison to the range of approaches that might be expected among physicians.
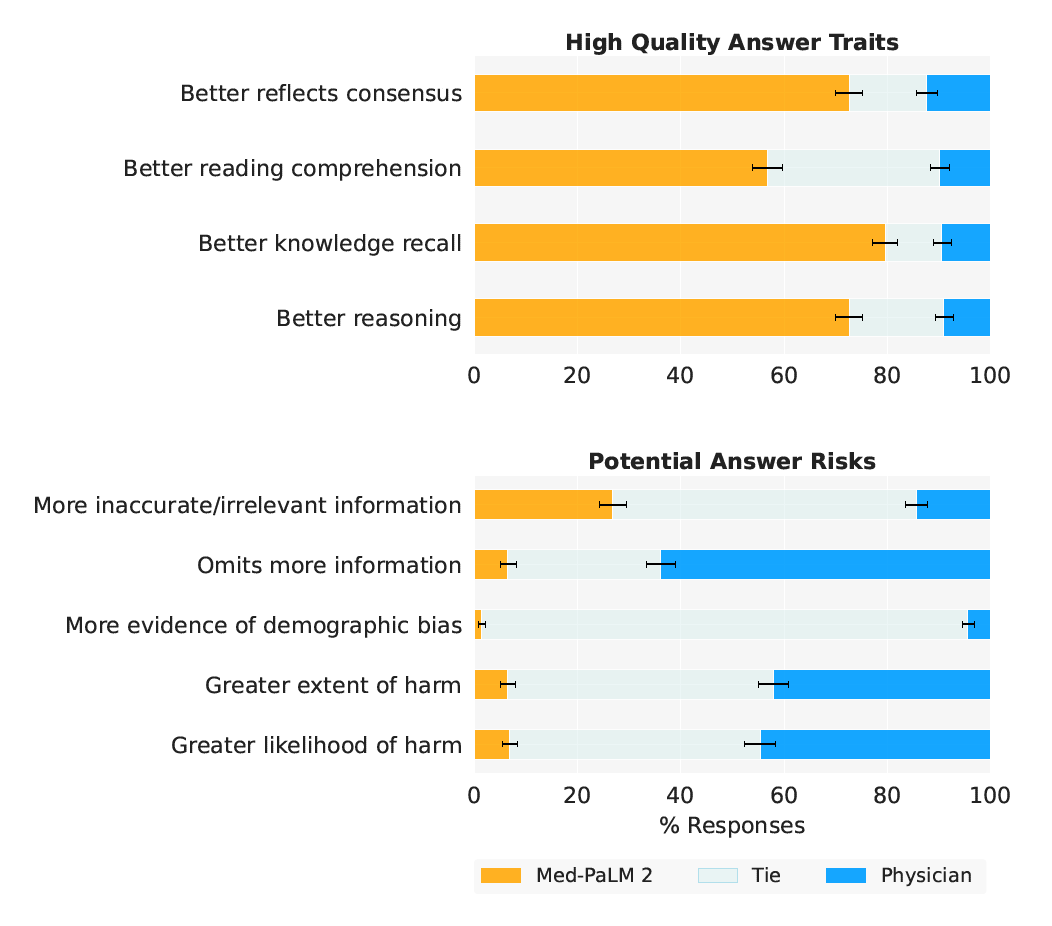
High Quality Answer Traits High Quality Answer Traits Better reflects consensus Better reflects consensus Better reading comprehension Better reading comprehension Better knowledge recall Better knowledge recall Better reasoning Better reasoning 0 20 40 60 80 100 0 20 40 60 80 100 . Potential Answer Risks Potential Answer Risks More inaccurate/irrelevant information More inaccurate/irrelevant information Omits more information Omits more information More evidence of demographic bias More evidence of demographic bias Greater extent of harm Greater extent of harm Greater likelihood of harm Greater likelihood of harm 0 20 40 60 80 100 % Responses 0 20 40 60 80 100 % Responses . (a) MultiMedQA (b) Adversarial Question Sets
- Figure 5 | Ranking comparison of long-form answers Med-PaLM 2 answers are consistently preferred over Med-PaLM answers by physician raters across all ratings dimensions, in both MultiMedQA and Adversarial question sets. Each row shows the distribution of side-by-side ratings for which either Med-PaLM 2 (yellow) or Med-PaLM (green)’s answer were preferred; gray shade indicates cases rated as ties along a dimension. Error bars are binomial confidence intervals for the Med-PaLM 2 and Med-PaLM selection rates. Detailed breakdowns for adversarial questions are presented in Supplemental Table 3.
Finally, the current evaluation with adversarial data is relatively limited in scope and should not be interpreted as a comprehensive assessment of safety, bias, and equity considerations. In future work, the adversarial data could be systematically expanded to increase coverage of health equity topics and facilitate disaggregated evaluation over sensitive characteristics [48–50] .
7 Conclusion
These results demonstrate the rapid progress LLMs are making towards physician-level medical question answering. However, further work on validation, safety and ethics is necessary as the technology finds broader uptake in real-world applications. Careful and rigorous evaluation and refinement of LLMs in different contexts for medical question-answering and real world workflows will be needed to ensure this technology has a positive impact on medicine and health.
...
References
- (Singhal et al., 2022) => Singhal, K., Azizi, S., Tu, T., Mahdavi, S. S., Wei, J., Chung, H. W., Scales, N., Tanwani, A., Cole-Lewis, H., Pfohl, S., et al. Large Language Models Encode Clinical Knowledge. arXiv preprint arXiv:2212.13138 (2022).
- (Nori et al., 2023) => Nori, H., King, N., McKinney, S. M., Carignan, D. & Horvitz, E. Capabilities of gpt-4 on medical challenge problems. arXiv preprint arXiv:2303.13375 (2023).
- (Liévin et al., 2022) => Liévin, V., Hother, C. E. & Winther, O. Can large language models reason about medical questions? arXiv preprint arXiv:2207.08143 (2022).
- (Google, 2023) => Google. PaLM 2 Technical Report https://ai.google/static/documents/palm2techreport.pdf. 2023.
- (Vaswani et al., 2017) => Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., Kaiser, Ł. & Polosukhin, I. Attention is all you need. Advances in Neural Information Processing Systems 30 (2017).
- (Devlin et al., 2018) => Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805 (2018).
- (Raffel et al., 2020) => Raffel, C., Shazeer, N., Roberts, A., Lee, K., Narang, S., Matena, M., Zhou, Y., Li, W. & Liu, P. J. Exploring the limits of transfer learning with a unified text-to-text transformer. The Journal of Machine Learning Research 21, 5485–5551 (2020).
- (Shortliffe, 1987) => Shortliffe, E. H. Computer programs to support clinical decision making. Jama 258, 61–66 (1987).
- (Schwartz, 1987) => Schwartz, W. B. Medicine and the computer: the promise and problems of change. Use and impact of computers in clinical medicine, 321–335 (1987).
- (Bobrow, 1994) => Bobrow, D. G. Categorical and probabilistic reasoning in medicine revisited (1994).
- (Yasunaga, Leskovec et al., 2022) ⇒ Yasunaga, M., Leskovec, J. & Liang, P. LinkBERT: Pretraining Language Models with Document Links. arXiv preprint arXiv:2203.15827 (2022).
- (Yasunaga, Bosselut et al., 2022) => Yasunaga, M., Bosselut, A., Ren, H., Zhang, X., Manning, C. D., Liang, P. & Leskovec, J. Deep bidirectional language- knowledge graph pretraining. arXiv preprint arXiv:2210.09338 (2022).
- (Bolton et al., 2022) ⇒ Bolton, E., Hall, D., Yasunaga, M., Lee, T., Manning, C. & Liang, P. Stanford CRFM Introduces PubMedGPT 2.7B https://hai.stanford.edu/news/stanford-crfm-introduces-pubmedgpt-27b. 2022.
- (Gu et al., 2021) => Gu, Y., Tinn, R., Cheng, H., Lucas, M., Usuyama, N., Liu, X., Naumann, T., Gao, J. & Poon, H. Domain-specific language model pretraining for biomedical natural language processing. ACM Transactions on Computing for Healthcare (HEALTH) 3, 1–23 (2021).
- (Luo, Sun et al., 2022) ⇒ Renqian Luo, Liai Sun, Yingce Xia, Tao Qin, Sheng Zhang, Hoifung Poon, and Tie-Yan Liu. (2022). “BioGPT: Generative Pre-trained Transformer for Biomedical Text Generation and Mining.” In: Briefings in Bioinformatics, 23(6). doi:10.1093/bib/bbac409
- Jin, D., Pan, E., Oufattole, N., Weng, W.-H., Fang, H. & Szolovits, P. What disease does this patient have? a large-scale open domain question answering dataset from medical exams. Applied Sciences 11, 6421 (2021).
- Pal, A., Umapathi, L. K. & Sankarasubbu, M. MedMCQA: A Large-scale Multi-Subject Multi-Choice Dataset for Medical domain Question Answering in Conference on Health, Inference, and Learning (2022), 248–260.
- Jin, Q., Dhingra, B., Liu, Z., Cohen, W. W. & Lu, X. PubMedQA: A dataset for biomedical research question answering. arXiv preprint arXiv:1909.06146 (2019).
- (Brown et al., 2020) ⇒ Brown, T., Mann, B., Ryder, N., Subbiah, M., Kaplan, J. D., Dhariwal, P., Neelakantan, A., Shyam, P., Sastry, G., Askell, A., et al. Language models are few-shot learners. Advances in Neural Information Processing Systems 33, 1877–1901 (2020).
- (Chowdhery et al., 2022) => Chowdhery, A., Narang, S., Devlin, J., Bosma, M., Mishra, G., Roberts, A., Barham, P., Chung, H. W., Sutton, C., Gehrmann, S., et al. PaLM: Scaling language modeling with pathways. arXiv preprint arXiv:2204.02311 (2022).
- Chung, H. W., Hou, L., Longpre, S., Zoph, B., Tay, Y., Fedus, W., Li, E., Wang, X., Dehghani, M., Brahma, S., et al. Scaling instruction-finetuned language models. arXiv preprint arXiv:2210.11416 (2022).
- Levine, D. M., Tuwani, R., Kompa, B., Varma, A., Finlayson, S. G., Mehrotra, A. & Beam, A. The Diagnostic and Triage Accuracy of the GPT-3 Artificial Intelligence Model. medRxiv, 2023–01 (2023).
- Duong, D. & Solomon, B. D. Analysis of large-language model versus human performance for genetics questions. medRxiv, 2023–01 (2023).
- Oh, N., Choi, G.-S. & Lee, W. Y. ChatGPT Goes to Operating Room: Evaluating GPT-4 Performance and Its Potential in Surgical Education and Training in the Era of Large Language Models. medRxiv, 2023–03 (2023).
- Antaki, F., Touma, S., Milad, D., El-Khoury, J. & Duval, R. Evaluating the performance of chatgpt in ophthalmology: An analysis of its successes and shortcomings. Ophthalmology Science, 100324 (2023).
- Ayers, J. W., Poliak, A., Dredze, M., Leas, E. C., Zhu, Z., Kelley, J. B., Faix, D. J., Goodman, A. M., Longhurst, C. A., Hogarth, M., et al. Comparing Physician and Artificial Intelligence Chatbot Responses to Patient Questions Posted to a Public Social Media Forum. JAMA Internal Medicine (2023).
- Wang, X., Wei, J., Schuurmans, D., Le, Q., Chi, E. & Zhou, D. Self-consistency improves chain of thought reasoning in language models. arXiv preprint arXiv:2203.11171 (2022).
- Sun, Z., Wang, X., Tay, Y., Yang, Y. & Zhou, D. Recitation-Augmented Language Models. arXiv preprint arXiv:2210.01296 (2022).
- Madaan, A., Tandon, N., Gupta, P., Hallinan, S., Gao, L., Wiegreffe, S., Alon, U., Dziri, N., Prabhumoye, S., Yang, Y., et al. Self-refine: Iterative refinement with self-feedback. arXiv preprint arXiv:2303.17651 (2023).
- (Nair et al., 2023) => Nair, V., Schumacher, E., Tso, G. & Kannan, A. DERA: Enhancing Large Language Model Completions with Dialog-Enabled Resolving Agents. arXiv preprint arXiv:2303.17071 (2023).
- Hendrycks, D., Burns, C., Basart, S., Zou, A., Mazeika, M., Song, D. & Steinhardt, J. Measuring massive multitask language understanding. arXiv preprint arXiv:2009.03300 (2020).
- Abacha, A. B., Agichtein, E., Pinter, Y. & Demner-Fushman, D. Overview of the medical question answering task at TREC 2017 LiveQA. in TREC (2017), 1–12.
- Abacha, A. B., Mrabet, Y., Sharp, M., Goodwin, T. R., Shooshan, S. E. & Demner-Fushman, D. Bridging the Gap Between Consumers’ Medication Questions and Trusted Answers. in MedInfo (2019), 25–29.
- Vyas, D. A., Eisenstein, L. G. & Jones, D. S. Hidden in plain sight—reconsidering the use of race correction in clinical algorithms 2020.
- Inker, L. A., Eneanya, N. D., Coresh, J., Tighiouart, H., Wang, D., Sang, Y., Crews, D. C., Doria, A., Estrella, M. M., Froissart, M., et al. New creatinine-and cystatin C–based equations to estimate GFR without race. New England Journal of Medicine 385, 1737–1749 (2021).
- Eneanya, N. D., Boulware, L., Tsai, J., Bruce, M. A., Ford, C. L., Harris, C., Morales, L. S., Ryan, M. J., Reese, P. P., Thorpe, R. J., et al. Health inequities and the inappropriate use of race in nephrology. Nature Reviews Nephrology 18, 84–94 (2022).
- Chen, I. Y., Pierson, E., Rose, S., Joshi, S., Ferryman, K. & Ghassemi, M. Ethical machine learning in healthcare. Annual review of biomedical data science 4, 123–144 (2021).
- Rigby, M. J. Ethical dimensions of using artificial intelligence in health care. AMA Journal of Ethics 21, 121–124 (2019).
- Williams, T., Szekendi, M., Pavkovic, S., Clevenger, W. & Cerese, J. The reliability of AHRQ Common Format Harm Scales in rating patient safety events. Journal of patient safety 11, 52–59 (2015).
- Williams, D. R., Lawrence, J. A., Davis, B. A. & Vu, C. Understanding how discrimination can affect health. Health services research 54, 1374–1388 (2019).
- Yearby, R. Structural racism and health disparities: Reconfiguring the social determinants of health framework to include the root cause. Journal of Law, Medicine & Ethics 48, 518–526 (2020).
- Wei, J., Wang, X., Schuurmans, D., Bosma, M., Chi, E., Le, Q. & Zhou, D. Chain of thought prompting elicits reasoning in large language models. arXiv preprint arXiv:2201.11903 (2022).
- Wang, b., Min, S., Deng, X., Shen, J., Wu, Y., Zettlemoyer, L. & Sun, H. Towards Understanding Chain-of-Thought Prompting: An Empirical Study of What Matters. arXiv preprint arXiv:2212.10001 (2022).
- Lewkowycz, A., Andreassen, A., Dohan, D., Dyer, E., Michalewski, H., Ramasesh, V., Slone, A., Anil, C., Schlag, I., Gutman-Solo, T., et al. Solving quantitative reasoning problems with language models. arXiv preprint arXiv:2206.14858 (2022).
- (OpenAI, 2023a) => OpenAI. GPT-4 Technical Report 2023. arXiv: 2303.08774 [cs.CL].
- Thoppilan, R., De Freitas, D., Hall, J., Shazeer, N., Kulshreshtha, A., Cheng, H.-T., Jin, A., Bos, T., Baker, L., Du, Y., et al. Lamda: Language models for dialog applications. arXiv preprint arXiv:2201.08239 (2022).
- Kossen, J., Cangea, C., Vértes, E., Jaegle, A., Patraucean, V., Ktena, I., Tomasev, N. & Belgrave, D. Active Acquisition for Multimodal Temporal Data: A Challenging Decision-Making Task. arXiv preprint arXiv:2211.05039 (2022).
- Weidinger, L., Mellor, J., Rauh, M., Griffin, C., Uesato, J., Huang, P.-S., Cheng, M., Glaese, M., Balle, B., Kasirzadeh, A., et al. Ethical and social risks of harm from language models. arXiv preprint arXiv:2112.04359 (2021).
- Liang, P., Bommasani, R., Lee, T., Tsipras, D., Soylu, D., Yasunaga, M., Zhang, Y., Narayanan, D., Wu, Y., Kumar, A., et al. Holistic evaluation of language models. arXiv preprint arXiv:2211.09110 (2022).
- Perez, E., Huang, S., Song, F., Cai, T., Ring, R., Aslanides, J., Glaese, A., McAleese, N. & Irving, G. Red teaming language models with language models. arXiv preprint arXiv:2202.03286 (2022).
;