Fimbriae
A Fimbria (plural Fimbriae) is a short filamentous structure that grows from the cytoplasmic membranes of Bacteria.
- AKA: Short Attachment Pilus, Attachment Pilus, Bacterial Fimbria.
- …
- Example(s):
- Counter-Example(s):
- a Conjugation Pilus,
- a Pilin.
- See: Bacteria, Microbiology. Mannose Receptor, Amyloid, Protein.
References
2018a
- (Wikipedia, 2018) ⇒ https://en.wikipedia.org/wiki/Fimbria_(bacteriology) Retrieved:2018-2-24.
- In bacteriology, a fimbria (plural fimbriae), also referred to as an "attachment pilus" by some scientists, is an appendage that can be found on many Gram-negative and some Gram-positive bacteria that is thinner and shorter than a flagellum. This appendage ranges from 3-10 nanometers in diameter and can be up to several micrometers long. Fimbriae are used by bacteria to adhere to one another and to adhere to animal cells and some inanimate objects. A bacterium can have as many as 1,000 fimbriae. Fimbriae are only visible with the use of an electron microscope. They may be straight or flexible.
Fimbriae carry adhesins which attach them to the substratum so that the bacteria can withstand shear forces and obtain nutrients. For example, E. coli uses them to attach to mannose receptors.
Some aerobic bacteria form a very thin layer at the surface of a broth culture. This layer, called a pellicle, consists of many aerobic bacteria that adhere to the surface by their fimbriae. Thus, fimbriae allow the aerobic bacteria to remain on the broth, from which they take nutrients, while they congregate near the air.
"Gram-negative bacteria assemble functional amyloid surface fibers called curli."[1] Curli are a type of fimbriae;[2] another type are called type I fimbriae. Curli are composed of proteins called curlins. Some of the genes involved are CsgA, CsgB, CsgC, CsgD, CsgE, CsgF, and CsgG.
- In bacteriology, a fimbria (plural fimbriae), also referred to as an "attachment pilus" by some scientists, is an appendage that can be found on many Gram-negative and some Gram-positive bacteria that is thinner and shorter than a flagellum. This appendage ranges from 3-10 nanometers in diameter and can be up to several micrometers long. Fimbriae are used by bacteria to adhere to one another and to adhere to animal cells and some inanimate objects. A bacterium can have as many as 1,000 fimbriae. Fimbriae are only visible with the use of an electron microscope. They may be straight or flexible.
- ↑ Epstein, EA; Reizian, MA; Chapman, MR (2009), "Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly.", J Bacteriol, 191 (2): 608–615, doi:10.1128/JB.01244-08, PMC 2620823 Freely accessible, PMID 19011034.
- ↑ Cookson, AL; Cooley, WA; Woodward, MJ (2002), "The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces", Int J Med Microbiol, 292 (3-4): 195–205, doi:10.1078/1438-4221-00203, PMID 12398210.
2018b =
- (Barbercheck et al., 2018) ⇒ Barbercheck, C. R. E., Bullitt, E., & Andersson, M. (2018). Bacterial Adhesion Pili. In Membrane Protein Complexes: Structure and Function (pp. 1-18). Springer, Singapore. PMID: 29464555 DOI: 10.1007/978-981-10-7757-9_1
- ABSTRACT: Escherichia coli bacterial cells produce multiple types of adhesion pili that mediate cell-cell and cell-host attachments. These pili (also called 'fimbriae') are large biopolymers that are comprised of subunits assembled via a sophisticated micro-machinery into helix-like structures that are anchored in the bacterial outer membrane. They are commonly essential for initiation of disease and thus provide a potential target for antibacterial prevention and treatment. To develop new therapeutics for disease prevention and treatment we need to understand the molecular mechanisms and the direct role of adhesion pili during pathogenesis. These helix-like pilus structures possess fascinating and unique biomechanical properties that have been thoroughly investigated using high-resolution imaging techniques, force spectroscopy and fluid flow chambers. In this chapter, we first discuss the structure of pili and the micro-machinery responsible for the assembly process. Thereafter, we present methods for measurement of the biomechanics of adhesion pili, including optical tweezers. Data demonstrate unique biomechanical properties of pili that allow bacteria to sustain binding during in vivo fluid shear forces. We thereafter summarize the current biomechanical findings related to adhesion pili and show that pili biomechanical properties are niche-specific. That is, the data suggest that there is an organ-specific adaptation of pili that facilitates infection of the bacteria's target tissue. Thus, pilus biophysical properties are an important part of Escherichia coli pathogenesis, allowing bacteria to overcome hydrodynamic challenges in diverse environments.
2017
- (Kaiser, 2017) ⇒ Gary Kaiser (2017). 2.5C: Fimbriae and Pili. In The Biology LibreTexts library.
- QUOTE: Fimbriae and pili are thin, protein tubes originating from the cytoplasmic membrane of many bacteria. Both are able to stick bacteria to surfaces, but pili are typically longer and fewer in number than fimbriae. They are found in virtually all Gram-negative bacteria but not in many Gram-positive bacteria. The fimbriae and pili have a shaft composed of a protein called pilin. At the end of the shaft is the adhesive tip structure having a shape corresponding to that of specific glycoprotein or glycolipid receptors on a host cell (Figure 2.5.1). There are two basic types of pili: short attachment pili and long conjugation pili.
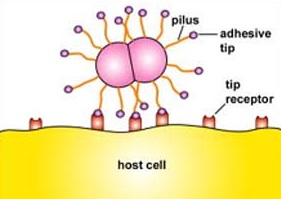
Figure 2.5.1: Adhesive Tip of Bacterial Pili Binding to Host Cell Receptors
Short attachment pili, also known as fimbriae, are usually short and quite numerous (Figure 2.5.1) and enable bacteria to colonize environmental surfaces or cells and resist flushing.
=== 2009 ===
- QUOTE: Fimbriae and pili are thin, protein tubes originating from the cytoplasmic membrane of many bacteria. Both are able to stick bacteria to surfaces, but pili are typically longer and fewer in number than fimbriae. They are found in virtually all Gram-negative bacteria but not in many Gram-positive bacteria. The fimbriae and pili have a shaft composed of a protein called pilin. At the end of the shaft is the adhesive tip structure having a shape corresponding to that of specific glycoprotein or glycolipid receptors on a host cell (Figure 2.5.1). There are two basic types of pili: short attachment pili and long conjugation pili.
- Gene Ontology http://amigo.geneontology.org/cgi-bin/amigo/term-details.cgi?term=GO:0009297&session_id=4446amigo1247242904
- Accession: GO:0009297
- Ontology: biological process
- Synonyms
- related: fimbria biogenesis
- related: fimbriae biogenesis
- related: fimbrium biogenesis
- related: pilus biogenesis
- narrow: fimbria assembly
- narrow: fimbriae assembly
- narrow: fimbrial assembly
- narrow: fimbrial biogenesis
- narrow: fimbrium assembly
- exact: pili biosynthesis
- exact: pili biosynthetic process
- exact: pilus biosynthesis
- exact: pilus formation
- Definition
- The assembly of a pilus, a short filamentous structure on a bacterial cell, flagella-like in structure and generally present in many copies. Pili are variously involved in transfer of nucleic acids, adherence to surfaces, and formation of pellicles. Is required for bacterial conjugation, or can play a role in adherence to surfaces (when it is called a fimbrium), and in the formation of pellicles. [source: GOC:dgh, GOC:mcc2, GOC:tb]
2007
- (Thanassi et al., 2007) ⇒ Thanassi D, Nuccio S, Shu Kin So S, Bäumler A. 2007. Fimbriae: Classification and Biochemistry, EcoSal Plus 2007; doi:10.1128/ecosalplus.2.4.2.1
- ABSTRACT: Proteinaceous, nonflagellar surface appendages constitute a variety of structures, including those known variably as fimbriae or pili. Constructed by distinct assembly pathways resulting in diverse morphologies, fimbriae have been described to mediate functions including adhesion, motility, and DNA transfer. As these structures can represent major diversifying elements among Escherichia and Salmonella isolates, multiple fimbrial classification schemes have been proposed and a number of mechanistic insights into fimbrial assembly and function have been made. Herein we describe the classifications and biochemistry of fimbriae assembled by the chaperone/usher, curli, and type IV pathways.
1993
- (Iriarte et al., 1993) ⇒ Iriarte, M., Vanooteghem, J. C., Delor, I., Diaz, R., Knutton, S., & Cornelis, G. R. (1993). The Myf fibrillae of Yersinia enterocolitica. Molecular microbiology, 9(3), 507-520. PMID 8105362
- ABSTRACT: “The Myf antigen produced by Yersinia enterocolitica appeared as a proteic polymer composed of 21 kDa subunits .By transposon mutagenesis we isolated Myf-defective mutants . Those allowed us to clone and sequence a 4.4 kb chromosomal locus involved in Myf production . This region was found to contain three genes that we called myfA, myfB and myfC .Genes myfB and myfC encode an assembly machine related to those involved in the synthesis of many fimbriae : MyfB, the putative chaperone, possesses the consensus residues of the PapD family and myfC encodes a putative outer-membrane protein .”
1991
- (Allen et al., 1991) ⇒ Allen, B. L., Gerlach, G. F., & Clegg, S. (1991). Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. Journal of bacteriology, 173(2), 916-920.PMID 1670938
- ABSTRACT: The nucleotide sequence of six genes involved in the expression of type 3 fimbriae of Klebsiella pneumoniae was determined. In addition to the genes that encode the fimbrial subunit (mrkA) and adhesion (mrkD), the mrkB, mrkC, and mrkE genes appear to be involved in assembly of the fimbrial filament and regulation of type 3 fimbrial expression. The mrkF gene product is required to maintain the stability of the fimbrial filament on the cell surface.