Named Entity Disambiguation (NED) Task
A Named Entity Disambiguation (NED) Task is a text processing task that requires mention linking in a text document with their correct referent entities in a knowledge base.
- AKA: Entity Linking Task, Named Entity Linking Task, Entity Disambiguation Task, Named Entity Recognition and Disambiguation Task, Named Entity Normalization Task, Named Entity Coreference Resolution Task.
- Context:
- Input:
- One or more Linguistic Expressions.
- One or more Entity Databases.
- optional: One or more Entity Types that the task is restricted to.
- output:
- A set of Entity Record Identifier for mentioned Entities.
- optional: A set of Entity Mention not present in the Entity Database.
- optional: A mapping from Entity Mention to Entity Record Identifier.
- It can be solved by an Named Entity Disambiguation System that applies an Named Entity Disambiguation Algorithm.
- It can range from being a Manual Named Entity Disambiguation Task to being an Automated Named Entity Disambiguation Task.
- It ranges from being an Unsupervised Named Entity Disambiguation Task to being a Supervised Named Entity Disambiguation Task.
- It can range from being a Data-Driven Entity Mention Linking Task to being a Heuristic Entity Mention Linking Task.
- It can be supported by an Entity Mention Coreference Resolution Task.
- Input:
- Example(s):
- a Text Wikification Task,
- a Disambiguation to Wikipedia (D2W) Task,
- a Collective Entity Linking Task,
- a Named Entity Mention Grounding Task,
- an Type-specific Entity Mention Normalization Task, such as:
- A Person Mention Normalization Task, such as: f("
[PERSON|Michael Jackson]is my favorite pop star") ⇒ “[PERSON_1734289|Michael Jackson] is my favorite pop star.". - A Gene Mention Normalization Task, such as a BioCreAtIvE II - Gene Normalization Task.
- A Product Mention Normalization Task.
- A Citation Mention Normalization Task.
- An Organism Mention Normalization Task.
- A Person Mention Normalization Task, such as: f("
- a Database-specific Entity Mention Normalization Task, such as:
- a Wikipedia-based Entity Mention Normalization Task that is restricted to linking to Wikipedia pages.
- a Gene Ontology-based Entity Mention Normalization Task that is restricted to linking to Gene Ontology ontology nodes.
- TAC-2014 Entity Linking Task.
- a Terminological Term Mention Linking Task.
- …
- Counter-Example(s):
- See: Semantic Search Task, Document To Entity Record Resolution Task, Entity Mention Coreference Resolution Task, Wikipedia, Knowledge.
References
2019
- (Wikipedia, 2019) ⇒ https://en.wikipedia.org/wiki/Entity_linking Retrieved:2019-6-16.
- In natural language processing, entity linking, named entity linking (NEL), named entity disambiguation (NED), named entity recognition and disambiguation (NERD) or named entity normalization (NEN) [1] is the task of determining the identity of entities mentioned in text. For example, given the sentence "Paris is the capital of France", the idea is to determine that "Paris" refers to the city of Paris and not to Paris Hilton or any other entity that could be referred as "Paris". NED is different from named entity recognition (NER) in that NER identifies the occurrence or mention of a named entity in text but it does not identify which specific entity it is.
Entity linking requires a knowledge base containing the entities to which entity mentions can be linked. A popular choice for entity linking on open domain text are knowledge-bases based on Wikipedia, [2] in which each page is regarded as a named entity. NED using Wikipedia entities has been also called wikification (see Wikify! an early entity linking system[3] ). A knowledge base may also be induced automatically from training text [4] or manually built.
Named entity mentions can be highly ambiguous; any entity linking method must address this inherent ambiguity. Various approaches to tackle this problem have been tried to date. In the seminal approach of Milne and Witten, supervised learning is employed using the anchor texts of Wikipedia entities as training data. [5] Other approaches also collected training data based on unambiguous synonyms. . Kulkarni et al. exploited the common property that topically coherent documents refer to entities belonging to strongly related types.
Entity linking has been used to improve the performance of information retrieval systems M. A. Khalid, V. Jijkoun and M. de Rijke (2008). The impact of named entity normalization on information retrieval for question answering. Proc. ECIR. </ref> and to improve search performance on digital libraries. [6] [7] NED is also a key input for Semantic Search. [8]
- In natural language processing, entity linking, named entity linking (NEL), named entity disambiguation (NED), named entity recognition and disambiguation (NERD) or named entity normalization (NEN) [1] is the task of determining the identity of entities mentioned in text. For example, given the sentence "Paris is the capital of France", the idea is to determine that "Paris" refers to the city of Paris and not to Paris Hilton or any other entity that could be referred as "Paris". NED is different from named entity recognition (NER) in that NER identifies the occurrence or mention of a named entity in text but it does not identify which specific entity it is.
- ↑ M. A. Khalid, V. Jijkoun and M. de Rijke (2008). The impact of named entity normalization on information retrieval for question answering. Proc. ECIR.
- ↑ Xianpei Han, Le Sun and Jun Zhao (2011). Collective entity linking in web text: a graph-based method. Proc. SIGIR.
- ↑ Rada Mihalcea and Andras Csomai (2007)Wikify! Linking Documents to Encyclopedic Knowledge. Proc. CIKM.
- ↑ Aaron M. Cohen (2005). Unsupervised gene/protein named entity normalization using automatically extracted dictionaries. Proc. ACL-ISMB Workshop on Linking Biological Literature, Ontologies and Databases: Mining Biological Semantics, pp. 17–24.
- ↑ David Milne and Ian H. Witten (2008). Learning to link with Wikipedia. Proc. CIKM.
- ↑ Hui Han, Hongyuan Zha, C. Lee Giles, "Name disambiguation in author citations using a K-way spectral clustering method," ACM/IEEE Joint Conference on Digital Libraries 2005 (JCDL 2005): 334-343, 2005
- ↑ [1]
- ↑ STICS
2013
- (Hachey et al., 2013) ⇒ Ben Hachey, Will Radford, Joel Nothman, Matthew Honnibal, and James R. Curran. (2013). “Evaluating Entity Linking with Wikipedia.” In: Artificial Intelligence, 194.
2011
- (Ji et al., 2011) ⇒ Heng Ji, and Ralph Grishman. (2011). “Knowledge Base Population: Successful Approaches and Challenges.” In: Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies.
- QUOTE: The main goal of KBP is to promote research in discovering facts about entities and augmenting a knowledge base (KB) with these facts. This is done through two tasks, Entity Linking -- linking names in context to entities in the KB -- and Slot Filling -- adding information about an entity to the KB.
2011a
- (Han et al., 2011) ⇒ Xianpei Han, Le Sun, and Jun Zhao. (2011). “Collective Entity Linking in Web Text: A Graph-based Method.” In: Proceedings of the 34th international ACM SIGIR conference on Research and development in Information Retrieval. doi:10.1145/2009916.2010019
- QUOTE: The key issue is to correctly link the name mentions in a document with their referent entities in the knowledge base, which is usually referred to as Entity Linking (EL for short). For example, in Figure 1 an entity linking system should link the name mentions “Bulls”, “Jordan” and “Space Jam” to their corresponding referent entities Chicago Bulls, Michael Jordan and Space Jam in the knowledge base.
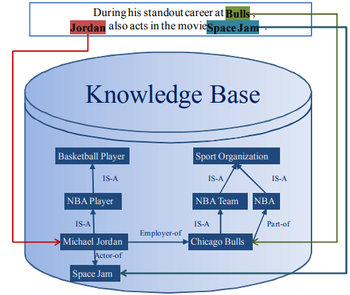
Figure 1. An illustration of entity linking.
- QUOTE: The key issue is to correctly link the name mentions in a document with their referent entities in the knowledge base, which is usually referred to as Entity Linking (EL for short). For example, in Figure 1 an entity linking system should link the name mentions “Bulls”, “Jordan” and “Space Jam” to their corresponding referent entities Chicago Bulls, Michael Jordan and Space Jam in the knowledge base.
2008a
- (Leitner et al., 2008) ⇒ Florian Leitner, Martin Krallinger, Carlos Rodriguez-Penagos, Jörg Hakenberg, Conrad Plake, Cheng-Ju Kuo, Chun-Nan Hsu, Richard Tzong-Han Tsai, Hsi-Chuan Hung William W Lau, Calvin A Johnson, Rune Sætre, Kazuhiro Yoshida, Yan Hua Chen, Sun Kim, Soo-Yong Shin, Byoung-Tak Zhang, William A. Baumgartner Jr, Lawrence Hunter, Barry Haddow, Michael Matthews, Xinglong Wang, Patrick Ruch, Frédéric Ehrler, Arzucan Özgür, Günes Erkan, Dragomir Radev, Michael Krauthammer, ThaiBinh Luong, Robert Hoffmann, Chris Sander, Alfonso Valencia. (2008). “Introducing Meta-Services for Biomedical Information Extraction.” In: Genome Biology, 9(Suppl 2):S6 doi:10.1186/gb-2008-9-s2-s6
- QUOTE: Entity mention normalization is based on large lexicon of known names and synonyms, which are kept in main memory at all times for efficiency. Once a potential named entity has been found, we further identify it using context profiles in case multiple entities share the same name [15];
Gene/protein normalization (GN): detect which genes or proteins are mentioned, assigning sequence database identifiers to the text.
The annotations we currently provide are gene mention normalization (32,795 human genes from EntrezGene)
We introduce the first meta-service for information extraction in molecular biology, the BioCreative MetaServer (BCMS; http://bcms.bioinfo.cnio.es/ webcite). This prototype platform is a joint effort of 13 research groups and provides automatically generated annotations for PubMed/Medline abstracts. Annotation types cover gene names, gene IDs, species, and protein-protein interactions. The annotations are distributed by the meta-server in both human and machine readable formats (HTML/XML). This service is intended to be used by biomedical researchers and database annotators, and in biomedical language processing. The platform allows direct comparison, unified access, and result aggregation of the annotations.
- QUOTE: Entity mention normalization is based on large lexicon of known names and synonyms, which are kept in main memory at all times for efficiency. Once a potential named entity has been found, we further identify it using context profiles in case multiple entities share the same name [15];
2008b
- (Morgan et al., 2008) ⇒ Alexander A Morgan, Zhiyong Lu, Xinglong Wang, Aaron M Cohen, Juliane Fluck, Patrick Ruch, Anna Divoli, Katrin Fundel, Robert Leaman, Jörg Hakenberg, Chengjie Sun, Heng-hui Liu, Rafael Torres, Michael Krauthammer, William W Lau, Hongfang Liu, Chun-Nan Hsu, Martijn Schuemie, K Bretonnel Cohen, and Lynette Hirschman. (2008). “Overview of BioCreative II gene normalization.” In: Genome Biology 2008, 9(Suppl 2):S3. doi:10.1186/gb-2008-9-s2-s3.
- QUOTE: The goal of the gene normalization task is to link genes or gene products mentioned in the literature to biological databases. This is a key step in an accurate search of the biological literature. It is a challenging task, even for the human expert; genes are often described rather than referred to by gene symbol and, confusingly, one gene name may refer to different genes (often from different organisms).
2008c
- (Farkas, 2008) ⇒ Richárd Farkas. (2008). “The strength of co-authorship in gene name disambiguation.” In: BMC Bioinformatics 2008, 9:69. doi:10.1186/1471-2105-9-69
- QUOTE: Taken one step further, the goal of Gene Name Normalisation (GN) [2] is to assign a unique identifier to each gene name found in a text.
2008d
- (JijkounKMR, 2008) ⇒ Valentin Jijkoun, Mahboob Alam Khalid, Maarten Marx, and Maarten de Rijke\n. (2008). “Named entity normalization in user generated content.” In: Proceedings of the second workshop on Analytics for Noisy Unstructured Text Data (AND 2008:23-30).
- QUOTE: Research on named entity extraction and normalization has been carried out in both restricted and open domains. For example, for the case of scientific articles on genomics, where gene and protein names can be both synonymous and ambiguous, Cohen [3] normalizes entities using dictionaries automatically extracted from gene databases.
2007
- (Morgan et al., 2007) ⇒ Alexander A. Morgan, Benjamin Wellner, Jeffrey B. Colombe, Robert Arens, Marc E. Colosimo, Lynette Hirschman. (2007). “Evaluating the Automatic Mapping of Human Gene and Protein Mentions to Unique Identifiers.” In: Pacific Symposium Biocomputing, 12.
- QUOTE:Vlachos et al. observed [19], in biomedical text there is a high occurrence of families of genes and proteins being mentioned by a single term such as: "Mxi1" belongs to the Mad (Mxi1) family of proteins, which function as potent antagonists of Myc oncoproteins". In future work in biomedical entity normalization, we suggest that normalizing entity mentions to family mentions may be an effective way to support other biomedical text mining tasks. Possibly the protein families in InterPro [6] could be used as normalization targets for mentions of families. For example, the mention of "Myc oncoproteins" could link to InterPro:IPR002418. This would enable information extraction systems that extract facts (relations, attributes) on gene families to attach those properties to all family members.
2006
- (Collier et al., 2006) ⇒ N. Collier, A. Nazarenko, R. Baud, and P. Ruch. (2006). “Recent advances in natural language processing for biomedical applications.” In: International Journal of Medical Informatics - Elsevier.
- QUOTE:... The BioCreative challenge on the other hand attempted to extend the traditional paradigm in two tasks that included entity mention normalization with respect ...
2005
- (Bretonnel Cohen et al., 2005) ⇒ K. Bretonnel Cohen, Lynette Hirschman, Hagit Shatkay, and Christian Blaschke. (2005). “Introduction to the Workshop on Linking Biological Literature, Ontologies and Databases: Mining Biological Semantics.” In: Proceedings of the ACL-ISMB 2005 Workshop on Linking Biological Literature, Ontologies and Databases: Mining Biological Semantics.
- QUOTE:In addition, we received submissions on the important topic of normalizing entity mentions.
2002
- (Cohen et al., 2002) ⇒ K. Bretonnel Cohen, Andrew Dolbey, George Acquaah-Mensah, Lawrence Hunter. (2002). “Contrast and Variability in Gene Names.” In: Proceedings of the ACL-2002 Workshop on Natural Language Processing in the Biomedical Domain. doi:10.3115/1118149.1118152
- QUOTE: Almost all current approaches to entity identification are actually not tackling the identification per se, but rather merely the (still difficult) location of named entities in text. The difference between these is that entity location consists of the (difficult enough) task of demarcation of the boundaries of names in text, whereas entity identification consists of the same thing, plus mapping the located names to the canonical entities that they refer to.
1992
- (Borgman & Siegfried, 1992) ⇒ C. L. Borgman, and S. L. Siegfried. (1992). “Getty's Synoname and Its Cousins: A survey of applications of personal name-matching algorithms.” In: Journal of the American Society for Information Science.