2016 TechMinerExtractingTechnologies
- (Osborne et al., 2016) ⇒ Francesco Osborne, Helene de Ribaupierre, and Enrico Motta. (2016). “TechMiner: Extracting Technologies from Academic Publications.” In: Proceedings of 20th International Conference on Knowledge Engineering and Knowledge Management (EKAW 2016).
Subject Headings: TechMiner System, Scholarly Ontology, Standard Scholarly Entity.
Notes
- It uses abstracts rather than the full text of publications (to test the value of the approach on a significant but manageable corpus).
Cited By
Quotes
Author Keywords
Abstract
In recent years we have seen the emergence of a variety of scholarly datasets. Typically these capture "standard" scholarly entities and their connections, such as authors, affiliations, venues, publications, citations, and others. However, as the repositories grow and the technology improves, researchers are adding new entities to these repositories to develop a richer model of the scholarly domain. In this paper, we introduce TechMiner, a new approach, which combines NLP, machine learning and semantic technologies, for mining technologies from research publications and generating an OWL ontology describing their relationships with other research entities. The resulting knowledge base can support a number of tasks, such as: richer semantic search, which can exploit the technology dimension to support better retrieval of publications; richer expert search; monitoring the emergence and impact of new technologies, both within and across scientific fields; studying the scholarly dynamics associated with the emergence of new technologies; and others. TechMiner was evaluated on a manually annotated gold standard and the results indicate that it significantly outperforms alternative NLP approaches and that its semantic features improve performance significantly with respect to both recall and precision.
1 Introduction
Exploring, classifying and extracting information from scholarly resources is a complex challenge and interesting challenge. The resulting knowledge base could in fact bring gamechanging advantages to a variety of fields: linking more effectively research and industry, supporting researchers’ work, fostering cross pollination of ideas and methods across different areas, driving research policies, and acting as a source of information for a variety of applications.
However, this knowledge is not easy to navigate and to process, since most publications are not in machine-readable format and are sometimes poorly classified. It is thus imperative to be able to translate the information contained in them in a free, open and machine-readable knowledge graph. Semantic Web technologies are the natural choice to represent this information and in recent years we have seen the development of many ontologies to describe scholarly data (e.g., SWRC1, BIBO2 , PROV-O3, AKT4) as well as bibliographic repositories in RDF [1, 2, 3]. However, these datasets capture mainly ‘standard’ research entities and their connections, such as authors, affiliations, venues, publications, citations, and others. Hence, in recent years there have also been a number of efforts, which have focused on extracting additional entities from scholarly contents. These approaches have focused especially on the biomedical field and address mainly the identification of scientific artefacts (e.g., genes [4], chemical components [5]) and epistemological concepts [6, 7, 8] (e.g., hypothesis, motivation, experiments). At the same time, the Linked Open Data cloud has emerged as an important knowledge base for supporting these methods [9, 10, 11].
In this paper, we contribute to this endeavour by focusing on the extraction of technologies, and in particular applications, systems, languages and formats in the Computer Science field. In fact, while technologies are an essential part of the Computer Science ecosystem, we still lack a comprehensive knowledge base describing them. Current solutions cover just a little part of the set of technologies presented in the literature. For example, DBpedia [12] includes only well-known technologies which address the Wikipedia notability guidelines, while the Resource Identification Initiative portal [13] contains mainly technologies from PubMed that were manually annotated by curators. Moreover, the technologies that are described by these knowledge bases are scarcely linked to other research entities (e.g., authors, topics, publications). For instance, DBpedia often uses relations such as dbp:genre and dct:subject to link technologies to related topics, but the quality of these links varies a lot and the topics are usually high-level. Nonetheless, identifying semantic relationships between technologies and other research entities could open a number of interesting possibilities, such as: richer semantic search, which can exploit the technology dimension to support better retrieval of publications; richer expert search; monitoring the emergence and impact of new technologies, both within and across scientific fields; studying the scholarly dynamics associated with the emergence of new technologies; and others. It can also support companies in the field of innovation brokering [14] and initiatives for encouraging software citations across disciplines such as the FORCE11 Software Citation Working Group5.
To address these issues, we have developed TechMiner (TM), a new approach which combines natural language processing (NLP), machine learning and semantic technologies to identify software technologies from research publications. In the resulting OWL representation, each technology is linked to a number of related research entities, such as the authors who introduced it and the relevant topics.
We evaluated TM on a manually annotated gold standard of 548 publications and 539 technologies and found that it improves significantly both precision and recall over alternative NLP approaches. In particular, the proposed semantic features significantly improve both recall and precision.
The rest of the paper is organized as follows. In section 2, we describe the TechMiner approach. In section 3 we evaluate the approach versus a number of alternative methods and in section 4 we present the most significant related work. In section 5 we summarize the main conclusions and outline future directions of research.
1 http://ontoware.org/swrc/ 2 http://bibliontology.com. 3 https://www.w3.org/TR/prov-o/ 4 http://www.aktors.org/publications/ontology 5 https://www.force11.org/group/software-citation-working-group
2 TechMiner
The TechMiner (TM) approach was created for automatically identifying technologies from a corpus of metadata about research publications and describing them semantically. It takes as input the IDs, the titles and the abstracts of a number of research papers in the Scopus dataset6 and a variety of knowledge bases (DBpedia [12], WordNet [15], the Klink-2 Computer Science ontology [16], and others) and returns an OWL ontology describing a number of technologies and their related research entities.
These include: 1) the authors who most published on it, 2) related research areas, 3) the publications in which they appear, and, optionally, 4) the team of authors who introduced the technology and 5) the URI of the related DBpedia entity. The input is usually composed by a set of publications about a certain topic (e.g., Semantic Web, Machine Learning), to retrieve all technologies in that field. However, TM can be used on any set of publications.
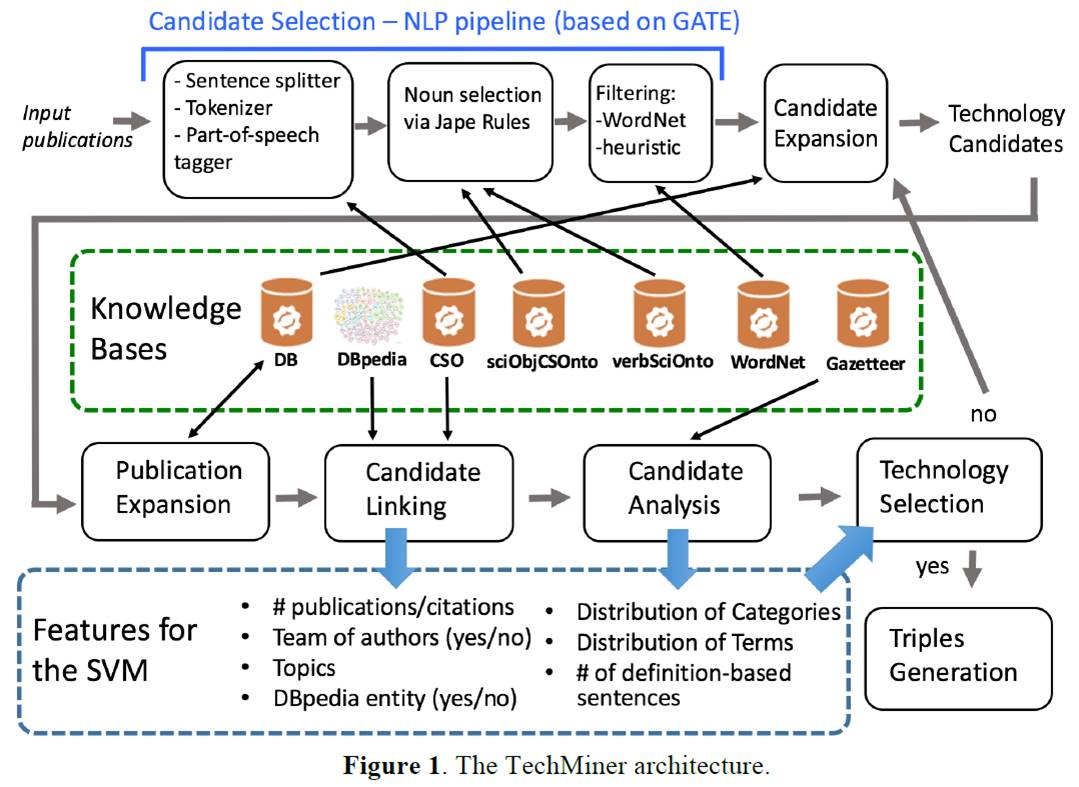
Figure 1. The TechMiner architecture.
We use abstracts rather than the full text of publications because we wanted to test the value of the approach on a significant but manageable corpus; in particular, one for which a gold standard could be created with limited resources. In addition, a preliminary analysis revealed that publications which introduce new technologies, a key target of our approach, typically mention them in the abstract.
6 https://www.elsevier.com/solutions/scopus
Figure 2 illustrates the architecture of the system, shows the adopted knowledge bases and lists the features that will be used by the classifier to detect if a candidate is a valid technology. The TM approach follows these steps:
- Candidate Selection (section 2.2). TM applies NLP techniques to extract from the abstracts a set of candidate technologies.
- Candidate Expansion (section 2.3). It expands the set of candidate technologies by including all the candidates discovered on different input datasets during previous runs which are linked to at least one of the input publications.
- Publication Expansion (section 2.4). It expands the set of publications linked to each candidate technology, using the candidate label and the research areas relevant to the associated publications.
- Candidate Linking (section 2.5). It applies statistical techniques to link each candidate to its related topics, authors and DBpedia entities.
- Candidate Analysis (section 2.6). It analyses the sentences in which the candidates appear and derives a weighted distribution of categories and terms.
- Technology Selection (section 2.7). It applies a support vector machine classifier for identifying valid technologies. If a candidate is not classified as a technology, TM returns to the Candidate Expansion phase, tries to further expand the set of publications linked to the candidate technology and repeats the analysis.
- Triple Generations (section 2.8). It produces the OWL ontology describing the inferred technologies by means of their characteristics and related entities.
In the next sections we shall discuss the background data and each step in details7.
2.1 Background data
For supporting the technology extraction task we manually crafted two ontologies: sciObjCSOnto8 and verbSciOnto9. The first was derived from sciObjOnto10 [17] and defines a number of categories of scientific objects in the Computer Science field and their related terms. It contains 47 classes/individuals, and 64 logical axioms and covers concepts such as: algorithm, application, software, implementation, model, approach and prototype. The verbSciOnto ontology was created to represent the verbs usually adopted for describing technologies (e.g., “describe”, “develop”, “implement”). It contains 26 classes and 67 individuals and 89 logical axioms. Each verb is described with its infinitive, past and present form.
In addition, TM exploits DBpedia, WordNet and the Klink-2 Computer Science Ontology. DBpedia is a well-known knowledge base, which derives from a community effort to extract structured information from Wikipedia and to make this information accessible on the Web. TM uses it to find entities associated to the candidate technologies, with the aim of yielding additional information for the technology extraction process. WordNet 11 is a large lexical database of the English language created by the Princeton University, and is widely used in the NLP field. TM exploits it to filter out generic nouns from the set of candidate technologies.
7 The ontologies, the JAPE rules and all the materials used for the evaluation is available at http://technologies.kmi.open.ac.uk/rexplore/ekaw2016/techminer/. 8 http://cui.unige.ch/~deribauh/Ontologies/sciObjCS.owl 9 http://cui.unige.ch/~deribauh/Ontologies/verbSciOnto.owl 10 http://cui.unige.ch/~deribauh/Ontologies/scientificObject.owl 11 https://wordnet.princeton.edu/wordnet/
The Klink-2 Computer Science Ontology (CSO) is a very large ontology of Computer Science that was created by running the Klink-2 algorithm [16] on about 16 million publications in the field of Computer Science extracted from the Scopus repository. The Klink-2 algorithm combines semantic technologies, machine learning and external sources to generate a fully populated ontology of research areas. It was built to support the Rexplore system (Osborne et al, 2013) and to enhance semantically a number of analytics and data mining algorithms. The current version of the CSO ontology includes 17,000 concepts and about 70,000 semantic relationships. The CSO data model12 is an extension of the BIBO ontology, which in turn builds on the SKOS model13. It includes three semantic relationships: relatedEquivalent, which indicates that two topics can be treated as equivalent for the purpose of exploring research data (e.g., Ontology Matching, Ontology Mapping), skos:broaderGeneric, which indicates that a topic is a sub-area of another one (e.g., Linked Data, Semantic Web), and contributesTo, which indicates that the research output of a topic contributes to another (e.g., Ontology Engineering, Semantic Web).
2.2 Candidate Selection
The aim of this first step is to identify a set of candidate technologies from an initial set of publications. To this end, TM processes the text of the abstracts by means of GATE[1], a well-known open source NLP platform, and a number of GATE plugins: OWLIM2, a module for importing ontologies, ANNIE, a component that forms a pipeline composed of a tokenizer, a gazetteer, a sentence splitter and a part-of-speech tagger, and JAPE (Java Annotation Patterns Engine), a grammar language for operating over annotations based on regular expressions.
The TM approach for identifying the set of candidates performs the following steps: 1) it splits the abstracts into sequences of tokens and assigns them part-of-speech tags (e.g., noun, verb and adverb) using ANNIE; 2) it selects technology candidates from sentences which contain a number of clue terms defined in the sciObjCSOnto ontology (e.g., “algorithm”, “tools”, “API”) and verbs from the verbSciOnto ontology (e.g., “implement”, “create”, “define” ) by applying a sequence of JAPE rules; 3) it filters the candidates by exploiting a number of heuristics.
A manual analysis on a variety of sentences about technologies revealed that the technology name can be a proper noun, a common noun or a compound noun, and not necessarily the subject or the object in the sentence. However, sentences about technologies are usually associated with a certain set of verbs and terms. For example, in the sentence: “DAML+OIL is an ontology language specifically designed for its use in the Web” the position of the noun “DAML+OIL” followed by the clue term “language” and subject of “is a”, suggests that DAML+OIL may be the name of a technology.
To identify similar occurrences, TM first uses 6 manually defined JAPE rules to detect a list of candidate nouns or compound nouns which cannot be authors, venues, journals or research topics. It then applies another set of 18 JAPE rules for identifying the sentences that contain both these candidate nouns and the clue terms from the sciObjCSOnto and verbSciOnto ontologies and for extracting the names of candidate technologies.
12 http://technologies.kmi.open.ac.uk/rexplore/ontologies/BiboExtension.owl 13 http://www.w3.org/2004/02/skos/
The rules were created following the methodology introduced in [17, 19] to construct JAPE rules from annotated examples. This approach clusters sentences that have similarities in the sequence of deterministic terms (e.g., terms and verbs described in the ontologies), then replaces these terms with either a JAPE macro or an ontology concept. Non-deterministic terms are instead replaced by a sequence of optional tokens.
In this instance, the rules were generated using examples from a dataset of 300 manually annotated publications from Microsoft Academic Search [19]. To improve the recall, we also created some additional JAPE rules to select also nouns that are not associated with any cue terms, but contain a number of syntactic grammatical patterns usually associated with the introduction of technologies.
The resulting candidates are then filtered using the following heuristics. We use WordNet to exclude common names by checking the number of synsets associated to each term contained in a candidate technology. A candidate associated with more than two synsets is considered a general term and gets discarded. However, we took in consideration some relevant exceptions. A preliminary analysis revealed in fact that a large number of technologies in the field of Computer Science are named after common nouns that belong to one or several categories of the Lexicographer Files of WordNet, such as animals (e.g., OWL, Magpie), artefacts (e.g., Crystal, Fedora) and food (e.g., Saffron, Java). Therefore, TM does not exclude the terms in these categories. In addition, we implemented two other heuristics. The first one checks if the term is capitalized or contains uppercase letters (e.g., Magpie, OIL, ebXML) and if so it preserves it even if WordNet suggests that it is a common name. The second one checks the terms that contain hyphens or underscore symbols. If both parts of the term are lower-case (e.g., task-based), they will be analysed separately by the WordNet heuristic, otherwise (e.g., OWL-DL, OWL-s) they will be considered as one word. The current prototype is able to process about 10,000 abstracts in one hour.
2.3 Candidate Expansion
The result of the previous phase is a set of candidate names linked to the publications from which they were extracted. However, the JAPE rules may have failed to recognize some valid technology which is actually mentioned in one of the input papers.
Nonetheless, the same technology may have been recognized in previous runs on a different set of initial papers. This happens frequently when examining datasets in different fields. For example, the application “Protégé”, may not be recognized when running on a Machine Learning dataset, since the few papers that would mention it may not have triggered the JAPE rules. However, if we already identified “Protégé” by previously analysing a Semantic Web dataset, we can exploit this knowledge to identify the instances of Protégé also in the Machine Learning dataset.
Therefore, in the candidate expansion phase TM enriches the set of candidates by including the technologies discovered during previous runs which were linked to one of the current input papers. This solution takes more time and can introduce some noise in the data, but it is usually able to significantly improve recall without damaging precision too much. We will discuss pros and cons of this solution in the evaluation section.
2.4 Publication Expansion
In this phase, we still may have missed a number of links between candidates and publications. In fact, the full Scopus dataset may have many other publications, not included in the initial dataset, that refer to the candidate technologies. It is thus useful to expand the set of links to collect more data for the subsequent analysis. TM does so by linking to a candidate technology all the papers in the Scopus dataset that mention the candidate label in the title or in the abstract and address the same research area of the set of publications associated to the candidate by the JAPE rules. In fact, taking into account the research area in addition to the label is useful to reduce the risk of confusing different technologies labelled with the same name. TM determines the research areas by extracting the full list of topics associated to the initial papers and finding the lowest common super topic which covers at least 75% of them according to the CSO ontology.
For example, given a candidate technology such as “LODifier”[9], TM will analyse the distribution of topics relevant to the associated papers and may find that most of them are subsumed by the Semantic Web topic, it will then associate the candidate with all the papers that contain the label “LODifier” and are tagged with “Semantic Web” or with one of its sub areas according to CSO, such as “Linked Data” and “RDF”.
Finally, the relationships between candidates and publications are saved in a knowledge base and can be used to enrich the set of candidates in the following runs.
This process is naturally less accurate than the NLP pipeline and can introduce some incorrect links. However, as discussed in the evaluation, the overall effect is positive since the abundance of links discovered in this phase fosters significantly the statistical methods used in the next steps.
2.5 Candidate Linking
In this phase, TM applies a number of heuristics to link the candidate to related research entities. In particular, it tries to link the candidate with 1) the team of authors who appear to have introduced the technology, 2) related concepts in the CSO ontology, and 3) related entities in DBpedia. The presence and quality of these links will be used as features to decide if the candidate is a valid technology. For example, the fact that a candidate seems to have been introduced by a well-defined team of researchers and is associated to a cohesive group of topics is usually a positive signal.
The authors who first introduce a technology tend to have the highest number of publications about it in the debut year and to be cited for these initial publications.
Hence, TM extracts the groups of authors associated to the candidate publications, merges the ones that share at least 50% of the papers, discards the ones who did not publish in the debut year, and assigns to each of them a score according to the formula:
??"#$%& = ()*+ ,$,_()*+ #)% (?? + 1 - ??????)89 :;<&* + #:,+ ,$,_#:,+ #)% (?? + 1 - ??????)89 :;<&* (1)
Here ??????: , ??????: , ??????_??????: , ??????_??????: are respectively the number of publications, citations, total publications (for all the papers associated to the candidate) and total citations in the ??-th year; ?????? is the year of debut of the candidate; ?????? is the current year and ? is a constant > 0 that modules the importance of each year (?=2 in the prototype). Since raw citations follow a power law distribution, we use instead the ratio of publications and citations [20]. Finally, we select the team associated with the highest score, but only if this is at least 25% higher than the second one. Therefore, only a portion of the technology candidates will be associated with an author’s team. Its presence will be used as binary feature in the classification process.
To identify the significant topics, TM extracts the list of keywords associated to the publications and infers from them a set of research areas in the CSO ontology. It does so by retrieving the concepts with the same label as the terms and adding also all their super-areas (the technique is implemented in the Rexplore system and discussed comprehensively in [18]). For example, the term “SPARQL” will trigger the homonym concepts SPARQL and subsequently super-topics such as RDF, Linked Data, Semantic Web and so on.
Finally, TM tries to link the candidate object with entities on DBpedia. It extracts all the sentences in the abstracts and titles which contain the candidate label and annotates them using DBpedia Spotlight [21]. The entity which is associated with at least 25% more instances than the others is selected as representative of the candidate. If this exists, TM links the candidate to this entity and saves the alternative names, the textual description in English (dbo:abstract), and a set of related entities via the dct:subject and rdf:type relations. The other entities annotated by DBpedia Spotlight will be used for the subsequent linguistic analysis.
2.6 Candidate Analysis
Intuitively a technology should be associated with a semantically consistent distribution of terms related to a specific context (e.g., “tool”, “web browser”, “plugin”, “javascript”). Learning these linguistic signs can help to detect a valid technology. The papers retrieved during the paper expansion phases and the entities retrieved by DBpedia should thus contain a good number of these kinds of terms. Hence, TM scans 1) the abstracts of all related papers, 2) the labels of the entities annotated by DBpedia Spotlight, and, if it exists, 3) the abstract of the linked DBpedia entity and the labels of its related entities for significant terms in an automatically created gazetteer of keywords related to technologies. The gazetteer was built by tokenizing the sentences associated to the annotated technologies in the gold standard from [19] and extracting the terms that were less than 5 tokens away from the technology names. We then removed stop words and selected the most frequent terms from this distribution, ending up with a gazetteer of 500 terms.
TM searches for the significant terms using five different techniques: 1) co-occurrence, in which it checks whether the terms occur in the same sentence as the candidate; 2) proximity-based, in which it checks whether the terms appear five words before or after a candidate; 3) definition-based, in which it checks whether each term t appears as part of a definition linguistic pattern, such as ‘X is a t’ or ‘t such as X’; 4) entity-based, in which it checks whether the terms appear as part of a linked DBpedia entity; 5) topicbased, in which it checks whether the terms appear in the related concepts of the CSO ontology. The result of this process is a distribution of terms, in which each term is associated with the number of times it co-occurred with the candidate label according to the different techniques. We then augment semantically these distributions by including all the concepts from the sciObjCSOnto ontology and assigning to them the total score of the terms which co-occur the most with each concepts label. For example, the category ‘application’ will co-occur the most with terms such as ‘applications’, ‘prototype’, ‘system’ and so on; hence, it will be assigned the sum of their scores.
The resulting distribution and the information collected in the previous phases are then used as features for selecting the valid technologies from the candidate group.
2.7 Technology Selection
All information collected in the previous phases is then used by TM to decide whether a candidate is a valid technology, by applying a support vector machine (SVM) classifier (adopting a radial basis function kernel) on the set of features extracted in section 2.4 and 2.5, representing both the linguistic signature of the associated papers and the related research entities. We take in consideration the following features (rescaled in the range [-1, 1]): i) number of publications and citations; ii) the presence of an associated team of authors (sec. 2.4, binary feature) ; iii) number of linked research areas in the first, second and third level of the CSO ontology (sec. 2.4); iv) presence of a DBpedia entity with the same label (sec. 2.4, binary feature); v) distribution of related categories and terms considering each of them as a distinct feature (sec. 2.5); vi) number of definition-based sentences addressing the candidate and one of the technology related terms (sec. 2.5).
When a candidate is classified as a technology, TM saves the related information and proceeds to analyse the next candidate, if it exists. When the candidate fails to be classified as a technology, TM tries to expand the candidate selection by using in the candidate expansion phase the super-topic of the previously high-level topic selected in the CSO. If there are multiple super topics, it selects the one associated with the lowest number of publications. For example, if the first topic was “Semantic Web”, the new one will be “Semantics”. The process ends when the candidate is classified as a technology, when the root ‘Computer Science’ is yielded, or after n failed attempts (n=2 in the prototype). The current prototype processes about 2,500 candidate technologies in one hour, taking in account also the queries to external sources (e.g., DBpedia).
2.8 Triple Generation
In this phase, TM generates the triples describing the identified technologies by associating each technology with: 1) the related papers, 2) the number of publications and citations, 3) the team of authors who introduced the technology, 4) the main authors, i.e., the 20 authors with most publications about the technology, 5) the main topics, i.e., the 20 most frequent topics, 6) the categories from sciObjCSOnto (associated with their frequency) and, possibly, 7) the equivalent DBpedia entity.
The output is a fully populated ontology of the technologies identified in the input dataset. To this end, we crafted the TechMiner OWL ontology15. Our intention was not to create ‘yet another ontology’ of the scholarly domain, but to craft a simple scheme for representing our output. For this reason we reused concepts and relationships from a number of well-known scholarly ontologies (including FABIO [22], FOAF16, CITO, SKOS, SRO17 , FRBR18 ) and introduced new entities and properties only when necessary. The main classes of the TechMiner OWL ontology are Technology, foaf:Person, to represent the researchers associated to the technology, Topic (equivalent to frbr:concept and skos:concept) and Category, representing the category of the technology (e.g., application, format, language).
15 http://cui.unige.ch/~deribauh/Ontologies/TechMiner.owl 16 www.xmlns.com/foaf/0.1/ 17 http://salt.semanticauthoring.org/ontologies/sro 18 http://purl.org/spar/frbr
3 Evaluation
We tested our approach on a gold standard (GS) of manually annotated publications in the field of the Semantic Web. To produce it, we first selected a number of publications tagged with keywords related to this field (e.g., ‘semantic web’, ‘linked data’, ‘RDF’) according to the CSO ontology. We then created an interface to annotate the abstracts with names and types of technologies. Since recognizing technologies in a field requires a certain degree of expertise, we asked a group of 8 Semantic Web experts (PhD students, postdocs, and research fellows) from The Open University and Oxford University to perform this task. In particular, we asked the annotators to focus on specific technologies which could be identified with a label, and not to consider very common ones, such as “web server”. Indeed, we wanted to focus on technologies used or introduced by researches that would usually not be covered by generic knowledge bases. To avoid typos or extremely uncommon labels, we discarded from the output the technologies with labels appearing only once in the full set of 16 million abstracts from the Scopus dataset of Computer Science. The resulting GS includes 548 publications, each of them annotated by at least two experts, and 539 technologies. In this evaluation we focus only on the identification of technologies, and not on the correctness of the links between the technology and other entities (e.g., authors), whose presence is simply used as features for the classification process and will be analysed in future work.
Our aim was to compare the performances of the different techniques discussed in this paper. In particular, we planned to assess the impact of the candidate linking and candidate analysis phases (section 2.5 and 2.6) versus the NLP pipeline, the effect of the semantic features introduced in section 2.6, and the impact of the candidate extension phase (section 2.3). Therefore, we compared the following approaches:
- NL: the classic NLP pipeline [19], as discussed in section 2.2, with no additional filters;
- NLW: the NLP pipeline which uses WordNet to discard generic terms;
- TMN: TM not using semantic features derived by linking the candidates to the knowledge bases (CSO, sciObjCSOnto, DBpedia) nor candidate expansion;
- TM: The full TM approach not using candidate expansion;
- TMN_E: TMN using candidate expansion;
- TM_E: The full TM approach using candidate expansion.
The last four approaches were trained using the gold standard from [19], which counts 300 manually annotated publications from Microsoft Academic Search. TMN_E and TM_E were then applied on a 3,000 publication sample (other than our GS) in the Semantic Web area and learned a total of 8,652 candidates, of which 1,264 were used during the evaluation run, being linked to the initial publications in the GS.
The evaluation was performed by running each approach on the abstracts of the 548 annotated publications in the GS. Since we intended to measure also how the popularity of a technology would affect the outcomes of the approaches, we performed six different tests with each method in which we considered only the technology labels which appeared at least 2, 5, 10, 20, 50 or 100 times in the full set of the Scopus dataset for Computer Science.
We intended to assess both 1) the ability of extracting the technologies from a set of publications, and 2) the ability of yielding a correct set of relationships between those technologies and the publications in which they are addressed. Hence, we computed recall and precision for both tasks. The significance of the results was assessed using non-parametric statistical tests for k correlated data: Wilcoxon's test for k=2 and Friedman's test for k>2.
Table 2 shows the performance of the approaches. We will first discuss the performance of the technology extraction task. The NL method is able to retrieve about half of the technologies with a precision of about 60%, when considering all labels. The introduction of the WordNet filter (NLW) improves significantly the precision (+12.7%, p=0.03), but loses some recall (-4.6%). TMN is able to further increase precision over NLW (+12.6%, p=0.03), lowering the recall to about 44%. The introduction of the semantic features (TM) improves both precision (+2.1%, p=0.03) and recall (+2.4%, p=0.03); in particular, TM obtains the best result among all approaches regarding precision (87.6%) and performs significantly better (p=0.03) than TMN, NLW and NL regarding F-measure.
The ability of TMN_E and TM_E to consider also pre-learnt candidates yields a massive increase in recall (respectively +38.2% and +38.8%), paying a relative small price in precision (-1.6% and -1.9%). Once again, the adoption of semantic features increases both precision and recall, yielding no apparent drawbacks. Hence, TM_E performs significantly better than TMN_E regarding F-measure (p=0.028). In general, TM_E outperforms all the other approaches for recall and F-measure (85.1%), being able to extract technologies with a recall of 84.2% and a precision of 86%.
Table 1. Precision and recall for the six runs of the six approaches. In bold the best result of each run.
The approaches that used only the NLP pipeline to identify the candidates (NL, NLW, TMN, TM) improved their recall when considering more popular labels, but also committed more errors. An analysis of the data reveals that this happens mainly because they identify as technologies other kinds of popular named entities (e.g., universities, projects) that, being associated with a great number of publications, have a large chance to be involved in some of the patterns that trigger the JAPE rules. The two solutions that enhance the candidate set (TMN_E, TM_E) suffer from the opposite problem; they tend to perform well when dealing with rare technologies with few occurrences, and not considering them lowers their recall.
Figure 2 shows the F-measure for all the approaches. TM_E yields the best performance (85.1% when processing all the technologies in the GS), followed by TMN_E, NLW, TMN and NL. The difference between the approaches is significant (p < 0.0001).
Figure 2. F-measure of the technology extraction task.
Figure 3. F-measure of the links between technologies and publications.
The results regarding the extraction of links between technologies and publications exhibit a very similar dynamic. As before, TM performs best in terms of precision and TM_E in terms of recall. The main difference is that in this test TM_E and TMN_E exhibit a lower precision. This is due to the fact that the method for linking pre-learnt candidates to publications is more prone to error that the NLP pipeline, which links only publications in which it finds a specific linguistic pattern. Figure 3 shows the Fmeasure regarding relationships. TM_E is again the best solution, followed by the other approaches in the same order as before. The difference among the methods is again highly significant (p< 0.0001).
In conclusion, the evaluation shows that the TM approach yields significantly better results than alternative NLP methods and that the introduction of semantic features further improves the performance. The use of pre-learnt candidates introduces a small amount of noise in the set of linked papers, but yields a important increase in recall.
4 Related Work
Extracting knowledge from the full text of research publications is an important challenge. A number of systems such as Microsoft Academic Search (academic.research.microsoft.com), Google Scholars (scholar.google.com), and others automatically extract the metadata of research publications and make them available online. The semantic web community contributed to this process by creating a number of scholarly repositories in RDF, such as Semantic Dog Food[1], RKBExplorer [2], Bio2RDF [3], and others.
A number of approaches apply named entity recognition and similar techniques for extracting additional information from the full text of research publications. These methods usually address the identification of scientific artefacts (e.g., genes [4], chemical [5]) and epistemological concepts [6] (e.g., hypothesis, motivation, background, experiment). For example, Groza [7] focused on the identification of conceptualization zones through a novel approach based on the deep dependency structure of the sentences. Ibekwe-Sanjuan and al [23] developed a methodology which combines surface NLP and Machine Learning techniques for identifying categories of information, such as objective, results, conclusion and so on. O'Seaghdha and Teufel [24] addressed instead the identification of the rhetorical zoning (based on argumentative zoning) using a Bayesian latent-variable model. The Dr. Inventor Framework [25] is a publicly available collection of scientific text mining components which can be used to support this kind of tasks.
TM can be classified under the first category, since technologies can be considered scientific objects. As in other methods crafted for this task, it uses a pipeline which includes NLP and machine learning; the main difference is that it focuses on technologies and introduces a number of new statistical and semantic techniques to foster the identification process.
The use of the Linked Open Data cloud for supporting named entity recognition has yielded good results. For example, the LODifier approach [9] combines deep semantic analysis, named entity recognition and word sense disambiguation to extract named entities and to convert them into an RDF representation. Similarly, the AGADISTIS [10] system is a knowledge-base-agnostic approach for named entity disambiguation which combines the Hypertext-Induced Topic Search algorithm with label expansion strategies and string similarity measures. However, this kind of systems can be used only for linking existing technologies to the related entities in knowledge bases, not for discovering new ones. Sateli and Witte [11] presented a method which combines NLP and named entity recognition based on the LOD cloud for identifying rhetorical entities and generating RDF triples describing them. Similarly to TM, they use GATE for NLP and DBpedia Spotlight [21] for linking terms in the publications to DBpedia entities.
However, TM uses a classifier to process a number of features derived from the linked research entities.
A number of agencies in the field of innovation brokering and ‘horizon scanning’ identify new technologies by manually scanning the web [14], leading to high costs and slow throughput. Automatic methods such as TM could bring a dramatic improvement in their workflow, by allowing the selection of a set of candidate technologies with high accuracy. The output produced by TM can also enrich a number of knowledge sources which index technologies, especially considering that, a good number of these, such as Google Patents, cover only patented technologies. As mentioned, DBpedia [12] also includes a number of well-known technologies, even if they are not always described thoroughly. Another interesting resource is the Resource Identification Initiative portal [13], an archive which collects and assigns IDs to a number of scientific objects, including applications, systems and prototypes.
5 Conclusions
We presented TechMiner, a novel approach combining NLP, machine learning and semantic technologies, which mines technologies from research publications and generates an OWL ontology describing their relationships with other research entities.
We evaluated TM on a gold standard of 548 publications and 539 technologies in the field of the Semantic Web. The evaluation showed that the use of semantic features significantly improves technology identification, and that the full hybrid method outperforms NLP approaches. These results suggest that using a combination of statistical information derived from the network of relevant of research entities (e.g., authors, topics) and background knowledge offers a competitive advantage in this task.
TM opens up many interesting directions of work. We plan to enrich the approach for identifying other categories of scientific objects, such as datasets, algorithms and so on. This would allow us to conduct a comprehensive study on the resulting technologies, with the aim of better understanding the processes that lead to the creation of successful technologies. We also intend to run our approach on a variety of other research fields and to this end we are testing some methodologies to automatically populate the supporting ontologies with terms automatically extracted from research papers [26]. Finally, since similar experiences in the field of biotechnology [13] highlighted the importance of manually curating this kind of data, we would like to build a pipeline for allowing human experts to correct and manage the information extracted by TechMiner.
References
- 1. Moller, K., Heath, T., Handschuh, S., Domingue, J.: "Recipes for Semantic Web Dog Food — The ESWC and ISWC Metadata Projects.” In: 6th International Semantic Web Conference, 11-15 Nov 2007, Busan, South Korea. (2007)
- 2. Glaser, H., Millard, I.: "Knowledge-Enabled Research Support: RKBExplorer.com.” Proceedings of Web Science 2009, Athens, Greece. (2009)
- 3. Dumontier, M., Callahan, A., Cruz-Toledo, J., Ansell, P., Emonet, V., Belleau, F., Droit, A.: "Bio2RDF release 3: a larger connected network of linked data for the life sciences.” International Semantic Web Conference (Posters & Demos) 2014. (2014)
- 4. Carpenter, B.: "LingPipe for 99.99% recall of gene mentions.” In: Proceedings of the Second BioCreative Challenge Evaluation Workshop (Vol. 23, pp. 307-309). (2007)
- 5. Corbett, P., Copestake, A.: "Cascaded classifiers for confidence-based chemical named entity recognition.” BMC bioinformatics, 9(11), p.1. (2008)
- 6. Liakata, M., Teufel, S., Siddharthan, A., Batchelor, C.R.: "Corpora for the Conceptualisation and Zoning of Scientific Papers.” In LREC. (2010)
- 7. Groza, T.: "Using Typed Dependencies to Study and Recognise Conceptualisation Zones in Biomedical Literature.” PloS one, 8(11), p.e79570. (2013)
- 8. de Ribaupierre, H., Falquet, G.: "September.” User-centric design and evaluation of a semantic annotation model for scientific documents. In: Proceedings of the 14th International Conference on Knowledge Technologies and Data-driven. (2014)
- 9. Augenstein, I., Padó, S., Rudolph, S.: "Lodifier: Generating linked data from unstructured text.” In The Semantic Web: Research and Applications (pp. 210-224). (2012)
- 10. Usbeck, R., Ngomo, A.C.N., Röder, M., Gerber, D., Coelho, S.A., Auer, S., Both, A.: "AGDISTIS-graph-based disambiguation of named entities using linked data.” In The Semantic Web–ISWC 2014 (pp. 457-471). Springer International Publishing. (2014)
- 11. Sateli, B., Witte, R.: "What's in this paper?: Combining Rhetorical Entities with Linked Open Data for Semantic Literature Querying.” In: Proceedings of the 24th International Conference on World Wide Web Companion (pp. 1023-1028). (2015)
- 12. Bizer, C., Lehmann, J., Kobilarov, G., Auer, S., Becker, C., Cyganiak, R., Hellmann, S.: "DBpedia-A crystallization point for the Web of Data.” Web Semantics: science, services and agents on the world wide web, 7(3), pp.154-165. (2009)
- 13. Bandrowski, A., Brush, M., Grethe, J.S., Haendel, M.A., Kennedy, D.N., Hill, S., Hof, P.R., Martone, M.E., Pols, M., Tan, S.C., Washington, N.: "The Resource Identification Initiative: A cultural shift in publishing.” Journal of Comparative Neurology, 524(1), pp.8-22. (2016)
- 14. Scanning Douw, K., Vondeling, H., Eskildsen, D., Simpson, S.: "Use of the Internet in scanning the horizon for new and emerging health technologies: a survey of agencies involved in horizon scanning.” J Med Internet Res, 5(1), p.e6. (2003)
- 15. Fellbaum, C.: "WordNet: An Electronic Lexical Database.” Cambridge, MA: MIT Press. (1998)
- 16. Osborne, F., Motta, E.: "Klink-2: integrating multiple web sources to generate semantic topic networks.” In The Semantic Web-ISWC 2015 (pp. 408-424). (2015)
- 17. de Ribaupierre, H., Falquet, G.: ", An Automated Annotation Process for the SciDocAnnot Scientific Document Model.” In: Proceedings of the fifth International Workshop on Semantic Digital Archives, TPDL 2015. (2015)
- 18. Osborne, F., Motta, E., Mulholland, P.: "Exploring scholarly data with Rexplore.” In The Semantic Web–ISWC 2013 (pp. 460-477). Springer Berlin Heidelberg. (2013)
- 19. de Ribaupierre, H., Osborne, F., Motta, E.: "Combining NLP and semantics for mining software technologies from research publications.” In: Proceedings of the 25th International Conference on World Wide Web (Companion Volume) (2016).
- 20. Huang, W.: "Do ABCs get more citations than XYZs? Economic Inquiry, 53(1), 773–789.” (2015)
- 21. Mendes, P.N., Jakob, M., García-Silva, A., Bizer, C.: "DBpedia spotlight: shedding light on the web of documents.” In: Proceedings of the 7th International Conference on Semantic Systems (pp. 1-8). ACM. (2011)
- 22. Peroni, S. and Shotton, D.: "FaBiO and CiTO: ontologies for describing bibliographic resources and citations.” Web Semantics: Science, Services and Agents on the World Wide Web, 17, pp.33-43. (2012)
- 23. Ibekwe-SanJuan, F. Fernandez S., Sanjuan E. and Charton E.: "Annotation of scientific summaries for information retrieval.” arXiv preprint arXiv:1110.5722. (2011)
- 24. O'Seaghdha, D., Teufel, S.: "Unsupervised learning of rhetorical structure with un-topic models.” In: Proceedings of the 25th International Conference on Computational Linguistics (COLING 2014). (2014)
- 25. Ronzano, F., Saggion, H.: "Dr.” Inventor Framework: Extracting Structured Information from Scientific Publications. Discovery Science (pp. 209-220). Springer International Publishing. (2015)
- 26. Bordea, G, Buitelaar, P. and Polajnar,T. Domain-independent term extraction through domain modelling, the 10th International Conference on Terminology and Artificial Intelligence (TIA 2013), Paris, France. (2013);
| Author | volume | Date Value | title | type | journal | titleUrl | doi | note | year | |
|---|---|---|---|---|---|---|---|---|---|---|
| 2016 TechMinerExtractingTechnologies | Enrico Motta Francesco Osborne Helene de Ribaupierre | TechMiner: Extracting Technologies from Academic Publications | 2016 |